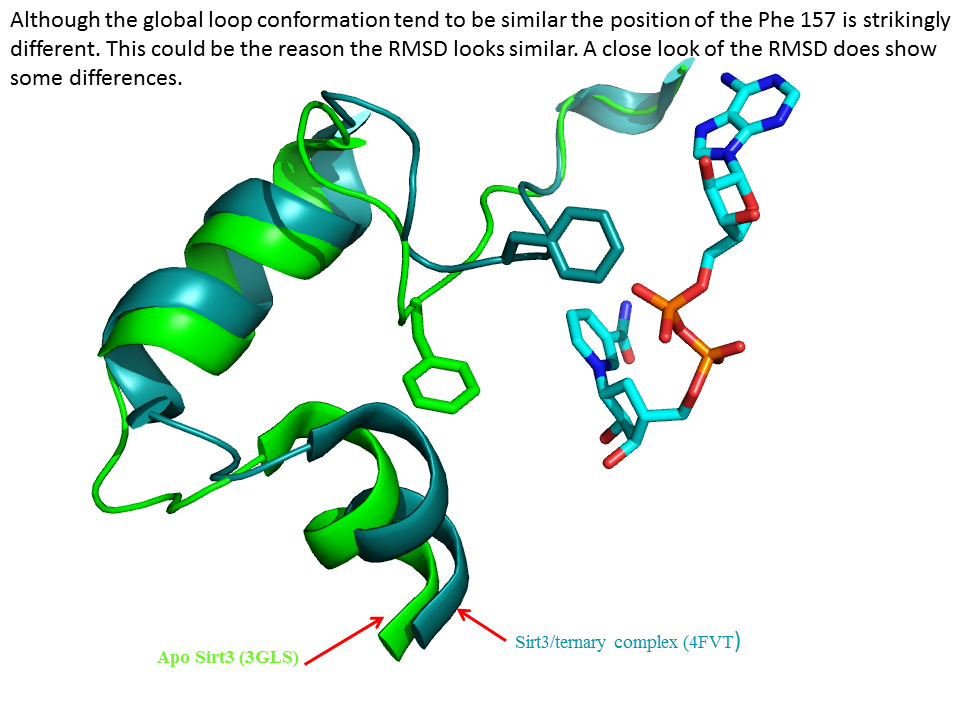
RSK: Schedule for Supplementary information
Supplementary_information_schedule.docx
RSK: Per-component/per-residue scores for open vs open and closed vs closed.
RSK: There are two sheetscomponent_score_per_reside_basis.xlsx
RSK: open vs open and closed vs closed
RSK: I analyzed the results of open vs open and closed vs closed. It looks like there are many residues which makes it hard to interpret when mapped onto the 3D coordinates. Anyways , I have the raw data.task_2_open_vs_open.xlsx
closedVSclosed.xlsx
RSK: Componenet score (prime) open vs open and closed vs closed
component_score_open_vs_prime.xlsxRSK: This contains data for open vs open ( ternary compared with product ) and closed vs closed ( ternary compared with product)
RSK: Results of task 2 and 4. The raw data will be uploaded to the dropbox.
RSK: Residues with significant delta E change between open and closed ( Delta E here is Native -non-native loop) . Those residues are mapped onto the 3D structureRSK:Report - This report describes the physical non-bonded change in the interactions in the residues when Sirt3 goes from an open to a closed conformation. This report will be updated for the product complex tonight.
RSK.The revised file is attched.
Report_open_vs_closed.docx
openVSclosed residue_merged.png.pptx
RSK Side chain prediction energies at various stages for xtal complex are provided. Sorry that I had the energy for the native 4BVG incorrect earlier.
Prime_side chain validation_xtal.xlsx
----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
RSK: Comparative plot of the per-residue energies for the product complex.
per_residue_product.pdfRSK: Revised scatter plot for 4FVT validation study. Prime side chain predicted vs Xtal and Prime side chain predicted with Prime Minimization vs Xtal
RSK: Will update the results for 4BVG shortly.RSK: It looks like that a global minimization on 4FVT would icrease the number of outliers ( wrong prediction).
RSK: Results for 4BVG
4BVG_new_plot.pdf
4FVT_comparasion_new.pdf
RK: Analysis to understand at which stage errors occurs, Its a elaborated study of task 2C.
RSK: Note here only native complexes were considered for validation. The results tend to indicate that errors could be due to energy function and sampling.modleled_vs_predicted.xlsx
RSK: Open vs closed MM/PBSA and GBSA energy comparison based on component terms for the complexes studied.
component_score_open_vs_close.xlsx
RSK: loop energies. It looks like the loop energies perform worse than complex energies.
loop_energy.xlsx
RSK: A report on "true outliers". The true outliers herein are those residues that appear as outliers in our validation study ( 4FVT and 4BVG) carried using crystal coordinates and cross comparing the same loop conformation when bound to different substrate/ligand( ie, ternary open vs Sirt3/INT/NAM open) .
RSK: Also, residues involved in substrate/ligand binding were excluded assuming that they could potentially have differenten bound to differentsubstrateeRSK: Task 2C Effect of Prime minimization. The data is attached . I will provide a short write up shortly. Also, the write up for per-residue energies will be provided today.
effect_of_minimi.xlsxRSK: Write up for 2C
2C_writeup.docx
RSK: Energy component analysis . First draft
first_draft_componentscore.docxRSK: I have analyzed the ternary open and closed complex based on the data which we have. Iam now working on the other sytems.
RSK: Please let me know if this is the kind of analaysis you are looking for.Ternarycomplex_report.docx
RSK: Per_residue energies for product complex (open and closed loop) and for ternary complex (closed loop). These are the three systems on which I performed MD simulations.I will now have to swithch to Amber 12 to computed the per-residue energies for SIRT3/INT/NAM and 4FVT ternary complex .These systems were handled by Ping and I belive that switcing over to Amber 12 form Amber 14 should fix the issue ( Although I am not certain ).
Per_residue_energies.xlsx
RSK: Per-residue energies report
RSK: Revised per-residue energies report along with decomposition and picture showing the crucial interactions.
per_residue_report.docx
RSK: Per-residue contribution for product complex (open and closed). A cut-off distance of 6A was used to compute the interactions.
Product_complex_4BVG.xlsx
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
RSK: Revised scatter plots . Red represents outliers (> 2A in RMSD and > +/- 4 kcal/mol ) . The residues are annotated ( Ligand binding, co-factor loop residues, other)
RSK: More aannotationswill be added as a note ( ie, the type of interaction )2A_new.pdf
------------------------------------------------------------
RSK: Component wise breakdo
wn for all the MD simulations. The shell script which I wrote for extracting them is also attached.
component_score.xlsxscore_extractor.sh
RSK: Attached is the summary of my analysis of 2A and 2B
2A_2B_Report.docx---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
RSK: Results for 2B
2B_Report.xlsxRSK: Scatter plot for 4FVT and 4BVG ( Red dots represents outlier)
Xtal_validation(RMSDvsDeltaE)_1.pdf
RSK: Scatter plot for 2A analysis ( Plots are named 2A and 2B) 2B name herein is not related to 2B analysis Task.
2A_correct_1.pdf
2B_plot_1.pdf
--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
RSK: Data for 2A and 4FVT pre/post validation, 4BVG pre/post validation. Contains delta E, RMSD and outlier residues
2A_@B_data.xlsx
RSK: 2A Report
Attched is an xls data sheet that contains the proceessed data for 2A analysis. (SIRT3/INT/NAM vs SIRT3/NAD+/AC-CS2 Open loop)Also a brief summary of the report. I belive that the plot and our analysis may need to be revised based on your feedback.
2A_Report.docx
2A_report.xlsx
---------------------------------------------------------------------------------
RSK: Revised work schedule.
Analysis Tasks_detailed workschedule_RC_comments_V2.docx-----------------------------------------------------------------------------------------------------------------------------------------------
RSK: Report based on analysis of our "consolidated MM/PBSA and MM/GBSA" data.
RSK:The edits are now contained in the "inconsitecy" document, corresponding to each section.
Inconsistencies in MM energy scoring 8-12_edited_RSK.docx
-------------------------------------------------------------------------------
RSK: The revised version of the schedule for analysis.
Analysis Tasks_detailed workschedule_RC_comments_V1.docx
--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
RSK: Detailed work schedule for the analysis part of the study.
Analysis Tasks_detailed workschedule.docxRC: Analysis Tasks_detailed workschedule_RC_comments.docx
Please note that after this schedule is settled we will need to add the remaining tasks that were on the original paper
completion schedule to see how long everything will take. Depending on the total time we may need to cut back on a few
analysis tasks for this paper.
RC (8/19): I look forward to your revisions to the schedule above per my comments today.
-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
RSK: I have uploaded Part I of the Knowledge transfer documents . Part I, here implies the data you requested for the Complex energies.Example files you requested will be separtely shared via dropbox later. A detailed document is provide. Please let me know if this letal of detailing is okay for the KT documents, if not I can revise it appropriately.
RSK: Also, I do have some queries as Iam not able to follow few of your comments here.Q1: 1E of the "Inconsistency of MM energy scoring" document has a comment "
"Note: we investigated rankings of apo structures because the complexes above were rank ordered incorrectly despite apparently correct rank ordering of ligand binding energies (see II below). Hence the protein conformational energies in the absence of ligand appear to be generally responsible for the inconsistencies" . Please advise me if my understanding here is correct or Iam not not really getting what you are trying to explain here.
To me it sounds that the "DERVIED Apo energies" comparasion may not be the ideal way to compare "receptor energies"
example
a) comparing ternary open vs ternary closed is not possible becuase one is an closed loop and the other is an open loop ( different conformation) altogether.
b) comparing ternary closed loop vs SIRT/INT/NAM closed loop is also not possible, because even here the recepror conformations are slightly different (ie, Sirt3 modelled in the presence of a ternary complex and sirt3 modelled in the presence of INT/NAM complex).
The prediction of side chains are largely influenced by the nature of the bound Ligand complex. Hence the derived Apo receptor energies may not be the actual "APO receptor energies" and may not be comparable between complexes with the same 4BVG/closed loop.
I may be wrong here please advise.
Q2 : I believe that we have not done any product complex simulation with 4BVH loop. ( I dont have data here)
Q3: I believe that we have not done any binary simulation with 4GLS loop. (I dont have data here)
RC: These queries have been addressed by email.
Knowledge_Transfer_PartI.docx
RC: Inconsistency of MM energy scoring
Inconsistencies in MM energy scoring 8-13.docxNote the magnitudes of the apparent scoring errors across many complexes.
Data still need to be added to this document. I hope this draft makes clear what has been requested.
I have included a comment regarding the fact that I need direct access to the data to avoid delays -- please review it and revert.
It may otherwise take many more weeks to make progress on this issue.
I have proposed detailed studies of various types for the purpose of analysis of the scoring errors in my recent emails.The order of work
has also been proposed.
If this is not clear shortly I may need to run the scripts myself.
The tasks listed in the document above are all in addition to other previously scheduled tasks needed for presentation of data in paper – need to consider the earliest those could finish given the new tasks and which new tasks need to be postponed to next paper
Will need estimated time for completion of each of these tasks
Note: if any specific MD simulations need to be re-run with modifications after the structure preparation errors are analyzed, these targeted simulations would need to run -while- the remaining paper tasks are completed.
SC has obtained a quote for a 2nd gpu node. Please consult on Mon regarding ordering of this node.
Knowledge transfer: start by providing RC with paths to all above data (including the filenames for each). RC must have direct access in case of delays.
Update: there are other knowledge transfer tasks described in the doc above.
email regarding apo receptor energies.docx
RSK:The revised versiosn is attcched
MM_GBSA_PBSA_consolidated_R1.xlsx
----------------------------------------------------------------------------------------------------------------------------------------------------------------
RSK: Consolidte data from Prime and Amber. This incudes Binding energies and decomposition values for the first frame (Prime/Amber) and ensembled avarged energies 2-12ns. Please note that I have now computed the input frame energy for 4FVT complex using Prime.
MM_GBSA_PBSA_consolidated.xlsx
RSK:
Energies after 2000 steps of Simple minimization in AmberRSK: This is in response to your earlier question
"Also, A,B) below were not both required -- I had indicated that you could use the easiest option so that consistent comparisons could be made across all pre-MD single point energies for the various complexes. If you did both for [A,B)] it is fine."
NB: Iam not able to locate the Intial frame and its corresponding topology file used by Ping. Hence, I am unable to calculate the energies (post 2000 steps of Amber minimization and the input frame here). However, we do have the MD trajectory. The first frame of the MD trajectory is after the relaxation phase.
Post_EM_Amber.xlsx
-------------------------------------------------------------------------------------
RSK: side chain validation data for all the complex are provided here. I have followed the same protocol used by ping on the native complex 4FVT ( ternary ) and 4BVG (Intermediate) complex.
RSK: Validation studies on two additrional systems were underataken by mea) Sirt3/INT/NAM moddlled on 4FVT
b) Sirt3/Product modelled on 4FVT
These, modelled complex were the ones for which side chain prediction was not carried out beforehand. I have now provided Prime energies pre and post side chain modelleing.
sidechain_validation.xlsx
RC: Ok I assume this provides comparable data for all complexes studied (with,without side chain prediction).
RSK: The data provide here is comparable as the same side chain prediction method has been employed across all complex.
RSK: The MM/GBSA energy compenets before and after side chain predcition are provided
sidechain_validation_energy_components.xlsx
RC: Ok by components I assume you mean complex, receptor, both ligands breakdown.
RSK: Thats right
RSK: If you look at the xls sheet the energy error for 4BVG ( Carried out by Ping) is approamiately -500 kcal. To me it raises a red flag because for all other systems the energy difference post side chain repacking is less that -50 kcal. There could be two issue shere
a) This is the only validation study we have on a 4BVG ( closed loop), so there is an possiplity that the diffrence in the oreintion of the closed loop (4BVG) could prove to be chalenge for Prime to recpatulate the exact orientaion of the side chains in this case
b) The value repored by ping could have been form an diffrent approcah.
Do you want me to reconfirm the numbers by repeating the validation study on 4BVG, so that we do have a confidence on the amount of noise propogated in an Model before ranking them.
RSK: I reran a calculation using 4BVG as a test case. it sounds that the energy error for 4BVG is ~500 kcal/mol.
-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
RSK: Attached is the revised protein-ligand interaction diagram. I have not considered NAM in these interaction plots (Because we can consider only one Ligand). Let me know if this representation is okay. If not we can revise it appropriately.Also, the peptide part of the INT molecule is excluded, to prevent the image being cluttered.
Ligplot_Interactions_Revised.pdf
RSK: I agree with your comments. Unfortunately, we don't have much control over these programs. They are mostly automated. However, I have created another version using the"poseview" program in-lieu of Schrodinger program.
Ligplot_Interactions_Revised_Poseview.pdf
-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
RSK: I have computed the Prime MM/GBSA and Amber MM/GBSA and MM/PBSA energies for the first frame ( ie, the Input structure for Amber ), Also I have reported the averaged 2-12 ns energies and break down numbers .
RC: Ok, to confirm: you have tabulated the consolidated xls with all the production simulation data including breakdown into complex, receptor, both ligands for all? If not, please do that.
And you have given this breakdown for first frames of all complexes as well, using the three scoring functions Prime MM/GBSA, Amber MM/GB and PBSA, except the one immediately below?
RSK: I have reported the production data results (2-12 ns) for
a) SIRT3/Ternary/4FVT loop (PLOS paper data)
b) SIRT3/Ternary/4BVG loop
c) SIRT3/INT/NAM (4FVT loop)
d) SIRT3/INT/NAM (4BVG loop)
This includes break down energies (Complex, Receptor, and Ligand), Prime MM/GBSA and Amber MM/GBSA and MM/PBSA energies for the complexes mentioned above.
Also, iam bit skeptical of the numbers for
RC: Ok, I believe I had flagged that as looking inconsistent as well. Please update that after your review.
RC: If the consolidated data is all in the xls entitled "first frame v3" please rename that accordingly so we know what is contained therein.
RSK: There is one issue here. I could not locate the INPUT frame for Sirt3/NAD+/Ac-CS ( 4FVT ) trajectory handled by Ping that is reported in the PLOS paper. The best guess we can do I, I can prepare an input from the crystal structure ( because no modelling is needed here ), just add hydrogen, and assume that he also used "protein prep" wizard of Schrodinger and then calculate the energy of the "input Frame",
RC: Not entirely clear why...you have the relaxation trajectory for that simulation? If so why not the input frame?
Related, you said you spent a day getting the Amber energies post initial minimization in the relaxation phase. Have you tabulated all of those?
If so, did you get it for the above complex as well?
I would need to have some comparison of an early frame for the ternary open loop complex. You can provide whatever is easiest.
RSK: I did mention that I have the energies post Initial minimization ie, the first step of the relaxation phase. I have also calculated the energies for all the complexes post 2000 steps of simple minimization. However, I have not tabulated them here assuming that you were not interested in those energies. Even in this case, I don’t have the values for Sirt3/NAD+/Ac-Cs (Ternary- 4FVT/PLOS). If needed I can post the values after 2000 step of minimization in Amber.
RSK: I just remember that Ping had a backup of most trajectories in the window machine also. I will check by logging in to the user account of Ping in the windows machine to see if it is lying there. I believe the initial PDB file he used as a starting point for simulation must have got deleted inadvertently. The first frame we have now in hand is the frame after post “Minimization” . If not I will create one on Monday and score it.
RSK: Secondly, I need your advice on comparing the Prime ( OPLS2005/VGSB2 ) with Amber . I got your point that we need to compare the "input frame" energies between Amber and OPLS 5 . Similarly, do we need to do such a compassion for the "LAST frame" ie 12 ns frame . We do have averaged values for 2-12 ns from Amber MM/PBSA and MM/GBSA values.
RC: Yes, we would like to do this, although first frame was most important since it related to the way the structures were prepared and we want to analyze those for consistency.
Ideally one could score the average structures with Prime, but I believe we said that could come later. I just want to mention again that for publication we may want to present that instead of just last frame.
I don't want to ask you to do last frame now for everything and then require you to do average for all again later (unless scoring last frame will be quite easy and fast for you to do).
Please consider what is best.
RSK: I am bit skeptical about computing energy values from time averaged structures, because the average structure which we compute from an MD trajectory tend to be non-physical (just an average of the Cartesian coordinates). These averaged structures had to be extensively energy minimized even to obtain meaningful geometries (bond length/torsion angles). Anyways I will do the last frame now as suggested. I will get this done on Monday
RC: I will review the latest data, but the ternary closed loop complex energy you provided earlier from Amber MD simulation was much lower than that of open loop, whereas this was not the case for INT/NAM. I need to check this for the first frames you are now posting. As a next step, we may structure align that for int/nam closed loop to that for ternary closed loop, and similarly for open loops. Backbones should be v similar. This could be a starting point for further analysis. You can proceed with this if you have the time now.
RSK: Okay, I will work on it. I will get you this data on Tuesday.
RSK: The requested structural; alignment figure is provided.
RSK. Attached is the revised figure with the right annotation
Closed Vs Open loop_starting frame.pdf
RC: Please confirm that neither of these involved any side chain prediction or MD, only minimization, starting from the same xtal structure.
RSK: The first pair ( Sirt3/Ternary- 4FVT loop compared with Sirt3/INT/NAM - 4FVT loop) doesn't involve any side chain prediction. However, the second pair does involve side chain prediction (Sirt3/Ternary - 4BVG loop compared with Sirt3/INT/NAM with 4BVG loop) . Only Prime energy refinement has been carried out.
RSK: I believe that Ping has not carried out any repacking/ loop refinement upon NAM placement.
RSK: looked at the side chain energies data which Ping has tabulated using 4FVT ( Ternary complex) as a model system for bench marking. Based on my inference of the data looking at the spread sheet, I could make out that he has calculated the "prime energy" after subjecting residues 155-178 ( ie, the co-factor loop region) to different "side chain prediction" methods as available in Prime/PLOP .
RC: Yes, isn't this quite analogous to how we prepared the complexes after loop substitution?
Hence I am comparing the reduction in energy due to side chain opt of a native complex to those after loop substitution.
It seems at first glance that the reduction in case of 4FVT native is much less than that observed for some of the loop substituted complexes (which you posted earlier).
Although anecdotal, this is good in that it doesn't suggest that side chain optimization of all native complexes would have reduced energy significantly.
RSK: The method is analogous; expect that we allowed more residues to be predicted. (Residues within 7.5 A of the loop region was allowed to be repacked). But now I have also reported the data for 4BVG. This is exactly similar to approach (loop + 7.5 A proximity repacked)
RC: Related, one interesting point to consider in analysis of the effect of side chain optimization (or lack thereof) in the rank ordering of open/closed loops is the fact that we ran int/nam simulations starting from both FVT and 4BVG protein structures. In the former case side chain optimization was applied to closed loop while in latter case side chain optimization was applied to open loop. In both cases during at least part of the simulation the closed loop complex had lower energy. Thus it is not observed that loop to which side chain optimization is applied always has lower energy. However note that a Phe flip was able to switch rank in latter case.
You can examine this simulation data from Ping if you have a chance.
RSK: That is a good point. May be we can considered plotting the RMSD of the Phe residue vs Potential Energy and see if they correlate. If it does, then we can convincingly state that switching of Phe trigger a shift in energy landscape. Let me know if you want me to do this.
What to do next with side chain prediction: I assume that only side chain prediction data you have on native loops is the aforementioned and posted data for 4FVT (none on 4BVG, e.g.).
We may want to continue to examine this effect of side chain prediction on native complex energies by applying the same protocol to all the complexes we have been simulating --
specifically, to the members of each pair (open or closed loop) to which side chain prediction was -not- previously applied before simulation (i.e., where the first frame was produced by
just minimization, no side chain prediction). Just tabulate the prime scores to start (complex). I want to check the magnitudes of the energy reduction due to side chain prediction and see
if it would have altered the rank ordering of loops.
This could be done relatively quickly, right?
After that, we could do further breakdowns/analyses if required.
RSK: I looked at the side chain energies data which Ping has tabulated using 4FVT ( Ternary complex) as a model system for bench marking. Based on my inference of the data from the spread sheet, I could make out that he has calculated the "prime energy" after subjecting residues 155-178 ( ie, the co-factor loop region) to different "side chain prediction" methods as available in Prime/PLOP . The four methods available in Prime are
A) Default—Use the default algorithm, which varies only the side chains and keeps the alpha and beta carbons fixed.
B) Monte Carlo—Perform Monte-Carlo sampling of side-chain conformations. When this algorithm is selected, the Maximum number of structures to return option and text box becomes available.
C) With CA-CB vector sampling—In addition to the default algorithm, vary the orientation of the CA-CB bond by up to 30°.
D) With backbone sampling—In addition to the default algorithm, perform a loop prediction on three residues: the residue whose side chain is being predicted and the residue on either side.
We are using option B and extending the repacking/prediction zone to 7.5A form the region of interest.
The energies are tabulate below.
Reference: Prime minimized = -12517.0
Default = -12493.2
CA-CB vector sampling = -12513.0
Monte Carlo MC (Rank 1) = -12528.8
Backbone sampling = -12530.1
I believe that during his benchmark study, residues with 7.5A was not considered for repacking, instead only residues 155-178 were repacked . That is what i infer looking at the xls sheet.
Also, attached is the raw data which I obtained from Ping's knowledge transfer documents. The number which I have reported above are form this document
4FVT-INT-NAM-s155-178-set1-DATA.xlsx
.
RSK: I was able to located the side chain validation data for 4BVG also from the Windows machine. This includes the loop region (Res 156-159 + 7.5 A proximity ).
Reference: Prime minimized = -9614.9
Default = -9896.1
CA-CB vector sampling = -10179.1
Monte Carlo MC (Rank 1) = -10175.5
Backbone sampling = -10268.2
The raw data file is also attached here.
full_RMSD_per_aa_4BVG-set1.xlsx
RSK: I have computed the Prime MM/GBSA and Amber MM/GBSA and MM/PBSA energies for the first frame ( ie, the Input structure for Amber ), Also I have reported the averaged 2-12 ns energies and break down numbers . The most time consuming part was preparing the system for Prime especially the INT/NAM complex which creates a lot of problem for reason I mentioned earlier. This caused me the delay.
RSK. There is one issue here. I could not locate the INPUT frame for Sirt3/NAD+/Ac-CS ( 4FVT ) trajectory handled by Ping that is reported in the PLOS paper. The best guess we can do I, I can prepare an input from the crystal structure ( because no modelling is needed here ), just add hydrogen, and assume that he also used "protein prep" wizard of Schrodinger and then calculate the energy of the "input Frame", However, this would take me at least half a day for setting up in Amber ( ie, adding the exact RESP charges for NAD+, parameter for Ac-Lys just like Ping did). Please let me know if this should be done.
RSK: Secondly, I need your advise on comparing the Prime ( OPLS2005/VGSB2 ) with Amber . I got your point that we need to compare the "input frame" energies between Amber and OPLS 5 . Similarly, do we need to do such a compassion for the "LAST frame" ie 12 ns frame . We do have averaged values for 2-12 ns from Amber MM/PBSA and MM/GBSA values.
firstframe_V3.xlsx
RC: If this is the latest 2-12 ns xls with all the complexes, as well as all the relevant input frame data with all energy functions, please rename accordingly and post it at the top of the page so all the latest data is collected
in one place for easy review. Please confirm that you are doing this. Thanks.
MM_GBSA_PBSA_consolidated.xlsx
RSK:
I will get the interaction plots today and will finish the script that automates the calculation of the Energy component as reported in the PLOS one paper.
Tomorrow, I will report the Amber MM/PBSA energy components in a format as shown in the PLOS paper ( I will also consolidate the 2-12 ns simulation data for all systems in to single xls sheet)
RC: Regarding consolidation of data, please note that the coproduct xls sheet below does not include receptor/ligand breakdown; when you consolidate please include those columns.
Moving on, I will report the prime energy breakdown values for the Prime MM/GBSA run
RC: See if you can consolidate data (esp for production runs) including breakdowns for all ligands in a single xls file (multiple sheets if required). There are a lot of spreadsheets posted below and the
only master spreadsheet I saw was for the 1 ns data.
RSK: General status
Protein-Ligand interaction plot completed ( Final version will be send out on Monday)
Rerunning the trajectories using the right salt con for MM/PBSA is complete ( done for all systems )
MM/GBSA and MM/PBSA energy components ( as per PLOS paper). perl script partially complete. Will be completed on Monday so that it can be automated and run across all systems of interest. Doing it manually will be more time consuming.
PENDING task
Prime energies break down values needs to be added
Time dependent RMSD for the acetyl oxygen distance 2- 12 ns ( one system complete , 4FVT ternary complex needs to be done) plot needs to be prepared .
Will prioritize my task for the next week based on your feedback.
RSK: First frame energies compared with 2- 12 ns average energies. Sirt3/ternary and Sirt3/INT complex in open (4FVT) and closed(4BVG) loop conformation are compared.
firstframe.xlsx
RSK: Attached is the first frame energies compared with averaged 2-12 ns time interval energies. The individual energy components are also provided. Pleas note that the energies of the SIRT3/INT/NAM complex have changed form the earlier version. The reason being, I realized today that the concatenated MD trajectory which Ping had has both the "Pre MD" and the "MD part", Hence, I reran them using the last fame of the energy minimized structure to make it consistent.
RC: What do you mean by "Pre MD" and "MD part" of MD trajectory?
When you say you reran them using last frame, which simulations/scoring calculations are you referring to?
firstframe_V1.xlsx
RSK: I meant the "Pre MD" part as the stage during which the protein is held fixed and the water molecules around it are allowed to equilibrate around and the system and the system being slowly heated form 0 to 300K. Once the temp of the system reaches 300K the protein side chains alone were subjected to 2000 steps of energy minimization ( the backbone is held fixed). The last frame of this step "Pre MD step" is taken forward for the "production MD". I used the last frame of the "Pre MD step" for computing the "First frame energy".
RC: I need to understand in detail what you have changed and why. Why is it relevant that the "concatenated MD trajectory" of Ping had both "Pre MD and MD parts" -- we already knew the MD protocol. When you said "
I reran them using the last fame of the energy minimized structure to make it consistent", consistent with what?
RSK: Consistent with what I was reporting for other systems ( First frame energy). I usually consider the "pre MD" part of the trajectory as a separate trajectory and I don't merge it with the MD trajectories obtained from different time interval to obtain a single trajectory. However, I realized lately that for the Sirt3/INT/NAM system, Ping has merged even the "Pre MD/ Equilibration phase" with the "MD part/Production phase". However, all his analysis are "correct" because his analysis has discarded the first 170 frames from the trajectory, which is right. Ideally I should have computed the energy at frame 170, instead of frame 0. That is what I did to correct it to make it consistent.
RC: This is a good opportunity to clarify the protocol:
a) the PLOS paper indicates the minimization occurs -before- a 200 ps MD simulation where main chain atoms are restrained. The slow heating up may have occurred before the 200 ps. This MD was referred to as relaxation phase. We may have been referring to it recently as equilibration (although we acknowledged that we would investigate convergence later).
b) as we recently discussed there was then a (unrestrained) phase that was not used for sampling, called the equilibration phase.
c) finally there was the sampling phase.
It appears in the INT/NAM simulations and our recent work as well, step b) was skipped in the sense that sampling started immediately after a).
Please confirm or revise. Please also indicate the total duration of a) -- 1 ns?
My original meaning when I said "first frame" and clarified "of equilibration" once you inquired, was to score after minimization, prior to any MD.
Since I already had the equilibration / relaxation phase scores for all complexes, defining first frame as occurring after that phase is not as useful.
I can review the results attached, but this may need to be revisited (how difficult would that be?)
RSK. Yes, its a good to get it clarified to ensure that we are on the same page. Sorry, if I sound to be verbose here.
Firstly the system is slowly heated form 0 to 300 K (Stage A) Once the system reaches 300 K a 2000 step energy minimization is done (to remove short contacts/ Steric bumps),this is stage B. Then a 200 ps restrained MD with the Backbone atoms restrained ( primarily to allow water molecules to diffuse into protein cavities) is done which is termed Stage C. Together, stages A to C is called as the equilibration phase/relaxation stage (PLOS terminology). The last frame from ( Stage C) is then used for an unrestrained production MD/ sampling phase. In the production MD as the system undergoes conformational sampling and we check for convergence during a production run. Basically the production run is broken down into a equilibrated phase and a pre-eqilbrated phase based on converge analysis.
RSK: The "First frame" energies which I am reporting are from Stage C, ie the last frame of the relaxation stage.
RSK: Pleas let me know if I have to "RE DO" it using the frame after the 200 step minimization. This is bit tricky and not straight forward , because, as I stated earlier Stages A to C are tied together in to a single trajectory/single phase and we need to identify that "particular frame" from that trajectory ( ie, the last frame after 2000 steps of minimization). I have also noted down your latest comments that in case if its needs to be re done it is NOT an priority right away.
Although I have reported the energies for NAD+ for 4FVT ternary complex data, I realized that I had not calculated the binding energies for Ac-CS peptide from Ping PLOS data.
I will report it once it get the values today.The MM/PBSA run was launched today afternoon. I expect it to be completed by 7 PM today. Once , I get the value for this this task would stand complete.
RC: Yes, we need the results for both INT and NAM ligands in the case of INT/NAM
and both NAD and peptide ligands in case of ternary.
RSK: The binding energies for INT, NAM, Ac-Pep and NAD+ are contained .
firstframe_V2.xlsx
RSK: Attached is a sample plot of the revise protein-ligand interaction. Let me know if this is fine, so that I can get it done for other systems also.
RMSD_plots_Revised_interactions_V3.pdf
RSK: Attached is the Ligand-Receptor interaction plot (2D plot) for Sirt3/INT/NAM with 4FVT and 4BVG loop and for Sirt3/ternary complex ( 4FVT and 4BVG loop). I am also working on creating Ligand -Recp interaction plots for the other systems too. Pleas let me know if you are okay with this interaction plot.
I didn't use Schrodinger for creating these plots, instead I used a free online server. Unfortunately, this server, don't provide us with any user parameter control ( ie, adjusting the non-bonded cut-off distance etc). All structures used herein are MD averages.
The link for the server is copied below. Please be noted here that this program is the one used by PDB for displaying protein-ligand interaction in complexes.
http://poseview.zbh.uni-hamburg.de/
Ligand _interactions.pdf
RSK: The revised RMSD and structural alignment figures are provided for the following pairs
Sirt3/INT/NAM (4BVG loop) vs Sirt3/NAD+/Ac-CS -4BVG loop or in other words ( CLOSED loop).
Sirt3/INT/NAM (4FVT loop) vs Sirt3/NAD+/Ac-CS -4BVG loop or in other words ( CLOSED loop).
and the other pair is
Sirt3/INT/NAM (4BVG loop) vs Sirt3/NAD+/Ac-CS -4FVT loop( native complex) or in other words ( OPEN loop).
Sirt3/INT/NAM (4FVT loop) vs Sirt3/NAD+/Ac-CS -4FVT loop( native complex) or in other words ( OPEN loop).
Th pairs of interest are highlighted in RED and GREEN to distinguish it from the other pairs studied here.
RMSD_plots_Revised.pdf
RSK: Thanks for clarifying. Regarding your concern regarding consistency, I think there is no issue, because I believe that I have always tagged the PDB ID to any model or calculations which I reported.
RSK: I confirm that ADPR/NAM complex is same as INT/NAM. I have reconciled this inconsistency in the revised table, which I will update shortly.
RSK: I now got it. You actually intended to compute the RMSD values for
Sirt3/INT/NAM (4BVG loop) vs Sirt3/NAD+/Ac-CS -4BVG loop or in other words ( CLOSED loop).
I have not done this, However, I see that I have reported the Sirt3/INT/NAM(4BVG loop) vs Sirt3/NAD+/Ac-CS -4FVT loop (OPEN loop)
RC: Yes, you are correct. Please bear in mind we would like to also see the structure alignment figure for this paper highlighting/zooming in on the ligands.
(It is good you did open loop as well so we can compare; if you have the structure alignment for that as well it will be useful.
From what I have seen so far it seems the RMSD will be quite a bit greater for the closed loop than for the open loop.)
The pending ligand interaction diagrams (please refer to prior correspondence for details), including annotation of the interatomic distance, will be relevant in this analysis as well.
Please let me know when you anticipate having each of the above.
RSK: MM/GBSA and MM/PBSA energies for Sirt3/Ac-CS/NAD+ with 4FVT loop ( Pings/PLOS data - simulation of native xtal structure ) vs Sirt3/AC-CS/NAD+ modeled based on 4BVG loop data is contained in the attached xls sheet. Note here that Pings PLOS trajectory was rerun using the latest version of the MMPBSA.py script from Amber for consistency reasons. The old values obtained for the complex energies and the new values are contained in the xls sheet. I am able to reproduce the binding affinity values reported in the PLOS paper. The need for re-running the trajectories arose because the complex energies calculated by Ping ( using the old script) was in orders of -24000 whereas the new script reports complex energies in the order of -7000 kcal/mol. However, the binding affinity values doesn't change between versions.
Hence if we need to compare the complex energies ( conformational energies) of Sirt3/Ac-CS/NAD+ 4FVT loop ( Pings/PLOS data) vs Sirt3/AC-CS/NAD+ modeled by me, we need to ensure consistency.
MMGBSA_plos_data_4BVG_ternary data_complex_V2.xlsx
RC (7-21): I have reviewed the complex energy data for open vs closed loops here, and it looks like there may be need to proceed with the analysis I suggested
earlier wherein we compare the energies of the first frames with the relevant energy functions. See my earlier comments and those dated 7-21 below.
Also, please confirm PLOS used 10-18 ns window for most of its reported scores.
RSK: Yes, looking at the raw data lying in the pmcatgpu1 node, its obvious that only if we consider the energies between 10-18 ns we are able to reproduce the number reported in PLOS paper.
RC: Why do you need to rerun two gbsa calcs. Please elaborate.
RSK: We need to re-run the Sirt3/ternary complex( 4FVT) trajectory reported in the PLOS paper using the latest version of the MMPBSA.py script for consistency reasons. The complex energies as per the old MMPBSA.py script are not comparable with the current version.
RSK: Secondly we need to re-score the Sirt3/INT/NAM trajectory done by Ping using the right salt concentration ( the PBSA part). Here we will have to run 4 calculations ie, Sirt3/INT/NAM with 4FVT loop using NAM as Ligand, Sirt3/INT/NAM with 4BVG loop using NAM as Ligand. Similarly, the same pair of calculations need to be done using Sirt3/INT/NAM with 4BVG loop.
RC: Regarding rmsd calcs note I was referring also to the structure alignment fig. Even if you did the relevant rmsd calc. Please read all my correspondence going back to
last week on this task carefully.
RSK: Sure .I believe that you are alluding to the below statement which I have copied below
RC: Regarding the requested RMSDs please see my comments in detail about this -- ADPR and NAM moieties from INT complex MD average (native closed loop) vs the respective moieties in NAD+ in the ternary complex MD average (closed loop in particular, also report open loop if you can conveniently get it or already have it -- I believe you may reported that before).
Going through the previous communications I see that the pending items are
Sirt3/Ternarycomplex (4FVT) MD average vs Sirt3/INT/NAM with 4FVT loop MD averaged
Sirt3/Ternarycomplex (4FVT) MD average vs Sirt3/INT/NAM with 4BVG loop MD averaged
This task includes corresponding structure alignment figures and reporting RMSD values for the entire complex, co-factor loop, ADPR part, NAM part and the peptide part with reference to the MD averaged structure of Sirt3/Ternary complex (4FVT)
RSK: The revised RMSD table and Structure alignment pictures. This includes
a) Sirt3/Ternarycomplex (4FVT) MD average vs Sirt3/INT/NAM with 4FVT loop MD averaged
b) Sirt3/Ternarycomplex (4FVT) MD average vs Sirt3/INT/NAM with 4BVG loop MD averaged
RMSD table_updated_3.docx
RMSC_plots.pdf
I have not considered the following systems for the time beginning
4BVG (native intermediate) MD average vs Sirt3/ADPR complex/NAM modelled from 4FVT (MD average)
4BVG (native intermediate) MD average vs Sirt3/ADPR complex/NAM modelled from 4FVT but with loop replaced form 4BVG (MD average)
I will update the RMSD for the above mentioned systems tomorrow. Please let me know if I have missed any systems from RMSD calculations.
RSK: As you stated in your email, I believe the confusion is regarding the loop terminologies. I guess that when you mean "closed loop" you are indicating the loop conformation seen in the ternary structure (Sirt3/Ac-ACS/NAD+ complex PDB ID: 4FVT). if that is the case then it has been already done.
RSK: I am sorry, going forward I will ensure that I will get things clarified if I am not clear before I proceed to work.
RSK: Update table(RMSD) and the structural alignments for Sirt3/INT/NAM with 4FVT loop compared with 4FVT and 4BVG xtal structures. Similarly Sirt3/INT/NAM with 4BVG loop compared with 4FVT and 4BVG xtal structures. Please let me know if these figures are okay, If needed I can revise them as required.
RMSD_plots.pdf
RMSD table_updated.docx
RSK: The 12 ns MM/GBSA and MM/PBSA values for Sirt3/NAD+/Ac-CS peptide (4BVG loop_. These values have been compared to the Sirt3/NAD+/Ac-CS peptide (4FVT) values reported in the PLOS paper.
RSK: I am able to reproduce the MM/GBSA and MM/PBSA values reported in PLOS paper.
Comparative data is contained in the attached xls
MMGBSA_plos_data_4BVG_ternary data.xlsx
RSK:I see that Ping has a file where there are complex energies for both MM/PBSA and MM/GBSA are there , however, it seems were obtained using an old version of the Amber MMPBSA.py script.
I think that for consistency reason will have to re-score the whole trajectory using the latest version of the Amber MMPBSA.py script ( which we used for all other trajectories). Please note here that the Binding affinity will not change between the versions however, the complex energy, receptor energy and the Ligand energy changes between version, differs. I will re score the trajectory using the latest version of MMPBSA.py script and send you the scores.
Anyways the complex energy form the old version is attached ( As reproduced form Pings data)
MMGBSA_plos_data_4BVG_ternary data_complex.xlsx
RSK: MM/GBSA and MM/PBSA results for Sirt3/ternarycomplex (4BVG loop)
MMGBSA_energies_4FVT_ternaray_4BVGloop.xlsx
RSK: Product complex 4FVT vs 4BVG loop MM/GBSA and MM/PBSA energies form 12 ns simulation
MMGBSA_12ns_energies_product_complex.xlsx
RSK: MM/GBSA and MM/PBSA results for Sirt3/ternarycomplex (4BVG loop)
MMGBSA_energies_4FVT_ternaray_4BVGloop.xlsx
The following are required before the results to date can be discussed.
--I have been reviewing the results for consistency.
-Related to the tasks scheduled for side chain prediction validation, we may need to consider issues with our application of side chain opt to one member of pair but not other. For now,I would like to see prime energies before/after side chain opt for complexes to which we applied side chain opt.
RSK: I have not looked at the side chain energies and I have dont have them handy. However, I am sure there are in the log file. I will extract these energies form the log file or project files.
RSK: I have compiled the data for 4FVT product complex with 4BVG loop (Sirt3/2'OAADPR/Deac-Pep/Intermediate loop) and 4FVT ternary complex with 4BVG loop (Sirt3/NAD+/Ac-CS2 peptide/Intermediate loop).
The Delta vales reported are the difference ( Side chain repacked - modeled structure) in energy of the side chains.
RSK: I have not considered the 4BVH system for the time being although we did side chain modelling on that system also.
RSK: Side chain for residues within 7.5 A of the grafted loop region (155-178) were repacked/remodeled using Prime followed by complete energy minimization using Prime.
Energy_values_4FVT_product_4BVGloop.xlsx
sidechain_eneregies_4FVT_ternary_4BVG.xlsx
RC (7-21): We will be able to use this data. However, I think there was a misunderstanding/communication on the particular task.
Here, I was looking for the total complex energy before and after side chain optimization.
The reason I am asking for this is that the side chain optimization was only carried out on closed loops as I recall (please indicate if not)
and this may "artificially" reduce the complex energies of closed loops but not open loops.
RSK(7/25): Side chain repacking/ remodeling was considered only when 4BVG loop ( closed form) was grafted onto 4FVT (ternary/ open ) crystal structures. The Prime energies of the complex pre and post side chain modelleing and refinement is provided below for the systems which I modeled.
4FVT_product complex with 4BVG loop = -11563.234 kcal/mol ( Before side chain modelling ie, Grafting of the loop followed by minimization of the complex using OPLS)
4FVT_product complex with 4BVG loop = -12213.082 kcal/mol (After side chain modeling ie, the best predicted side chain model followed by minimization of the complex using OPLS)
4FVT_ternary complex_4BVG loop = -11361.99 kcal/mol ( Before side chain modelling ie, Grafting of the loop followed by minimization of the complex using OPLS)
4FVT_ternary complex_4BVG loop = -12315.09 (After side chain modeling ie, the best predicted side chain model followed by minimization of the complex using OPLS)
In other words the energy which I have reported here is essentially the sum of all per-residue energies which I reported earlier in the xlsx sheet. However, the xlsx sheet doesn't contain the energies for the non-standard residues. The current values reported herein are the total energy of the complex (it includes non standard residues, Zn+, Ligand molecule etc.)
RC: Please post the numbers for INT/NAM complex above as well.
RSK: I don’t have the data handy, but I can check the files from Pings earlier work. I see that there is an xls sheet with some side chain energy data and RMSD. However, it’s more focused on comparing the side chain energies using different side chain modeling methods.
RC (7/26): Note that there is a significant energy change upon side chain optimization that is similar to or greater in magnitude to the complex energy differences for open and closed loops for the various complexes listed in the xls spreadsheets above.
This is partly due to inclusion of many side chains in the optimization. This suggests that we may need to subject the native structures to side chain optimization before MD as well to enable a proper comparison if we want to compare complex energies.
(We didn't initially do this to avoid altering the native structure due to energy or sampling errors in side chain prediction.) Alternatively, we may choose not to compare complex energies for open/closed loops,rather preferring to use the native structure without energy/sampling errors when it is available for one complex in the pair. We will need to discuss this.
RSK: I believe that comparing native ternary (4FVT) xtal structure with side chain modeled ternary (4FVT) with native intermediate (4BVG) xtal with side chain modeledintermediate will be a more realistic approach. We can estimate the conformational energy difference between modeled vs native ternary complex (4FVT) and modeled vs native intermediate (4BVG), and this difference should roughly be the equivalent of "energy error" form side chain modelling . However, this would require two more additional simulation involving modeling.
Before we can do so, we need to look at the corresponding energies for a native structure (no loop substitution) subject to analogous side chain prediction. This should have been done as part of the validation test set we have discussed many times in the past.
Please post such an example once you find it (e.g., 4FVT ternary complex with native loop, before/after side chain optimization).
RSK: I believe that we have not done any study comparing modeled side chain energies/RMSD with reference to native side chain energies from 4FVT or 4BVG as a model system for validation. However, Ping may have carried out studies along those lines, because I realize that he has carried some benchmark/validation exercise on side chain prediction accuracy. If needed, I can carry out a validation study to get an estimate of deviation in energies/RMSD for every amino acid when modeled. Ie Modeled vs Native energies for amino acids that were repacked/modeled and RMSD for every modeled amino acid with reference to its native structure.
This is related to the task I had assigned of rescoring the very first frame used in the various MD simulations with the relevant energy functions (in particular, Amber), esp for the INT/NAM and ternary complexes with open vs closed loops.
RC (7/26): See this comment as it affects our 7-26 correspondence on Amber scoring of "first frame". It should be the same structure as above, scored with Amber or prime (but broken down into complex,receptor,ligand).
The data above with OPLS seem to suggest that the side chain optimization on only one structure is confounding any complex energy comparisons (for OPLS, the relevant comparison is within an open/closed loop pair to determine whether OPLS/VSGB can properly rank order the two loops). The Amber first frame results (which rescore the same complexes) may lead to the same conclusion.
If you would like to discuss soon let me know, given that we may discuss the possibility of re-running some of the MD simulations after side chain optimization; I think it would be best to have the data requested above posted, though.
RSK: I agree that side chain optimization followed by energy optimization tends to push the structure to a minimum energy. More over since we had predicted the side chain rotamer of many residues here, it opens up the possibilities for errors, due to the combinatorial nature of the prediction and the pairwise additive approximation of the energy function involved.
RSK: I will get you the Amber scores after the 2000 steps minimization stage. As you stated the Amber MM/PBSA and MM/GBSA score may also point along the directions which Prime/MM-GBSA does.
Do you think that we should consider two more MD simulations on native complexes 94FVT and 4BVG) post side chain prediction/modelling ( ie loop region and 7.5 A within loop)
--MD average complex energies (Amber MM-GBSA) were:
INT/NAM open loop: -7146 closed loop: -7202. Closed is native structure in absence of NAM.
ternary open loop (2-18 ns?): -7355 closed loop: -7720. Open is native structure.
As noted I would like to compare the energies given that we have claimed the use of identical structure preparation methods for each. Please report the full breakdown (complex, receptor, ligands -- two if possible).
I need to verify the consistencies of the structure preparations to whatever extent possible before drawing conclusions that compare the scores.
A particular focus is the comparison of first frame scores for INT/NAM open and closed loop complexes, and also the comparison of first frame scores for ternary open and closed loop complexes.
It may be convenient to include these scores in the same spreadsheets as MD.
Please provide me a status update on this task. You can review the original assignment carefully first. I believe you may have done part but not all of it.
RC (7-22): Please let me know if you would be able to provide these today.
RSK: I can provide you the energies for the first frame before the end of the day( Amber MM/GBSA and MM/PBSA energies) . Also, when you imply here as the first frame, do you mean the First frame in the production run or the Equlibration run?
Also, please provide an update on prime rescorings.
These tasks are prerequisites for productive discussion of next steps as noted earlier.
-Prime may have an energy error for INT/NAM complex energies (open vs closed loop)
Need prime scores for both open/closed ternary loops before more complete analysis can be done.
RSK:Okay
RSK: I have the file which I got it from Pings data. However, I don't see that the energy values for every residue to be reported there ( only the total energy is reported). Its sounds that his analysis was more focused on RMSD . Although not sure which was the reference state for his RMSD calculation. I have attached the data file which i got form Ping folder
RC: Not sure we're on the same page here. I just mean that we need to score the ternary complexes (starting structures used for open and closed loop simulations) with prime and compare the scores. I believe you said these were pending.
RSK: I got it now. I misunderstood that you want to have a look at the Side chain energies for these models too..
4FVT-INT-NAM-set1-ref-4BVG.xlsx
--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
Need receptor, ligand breakdowns for all complexes (I believe you are providing it for Amber). Might include full breakdown for prime as well (less imp).
RSK: Yes, I have been reporting them for Amber MM/GBSA and MM/PBSA
-Please double check the breakdown for coproduct open loop. Upon a quick review it appeared there may be a typo in receptor or complex energy
RSK: I see, I will check it for sure. Sorry about that
-In order to further investigate consistency (esp noting that some of the simulations were set up by Ping and others by you) I would like to compare receptor and binding energies for first frame of INT/NAM sims (open and closed loops) to the respective ternary sims – ideally with all energy fns. If prime is easiest, you can report that to start. Use the same initial structures used for MD simulations in each case.
-- Regarding geometries from the closed loop ternary complex simulation, provide RMSDs of ADPR moiety and NAM moiety in the closed loop xtal structure to the respective moieties in NAD from the closed loop ternary complex simulation. Also show the structure alignment. Related, update “SI Table …….: Table showing RMSD comparison between various Sirt3 complexes” with the latest simulation data.
--Please send annotated ligand interaction diagrams, scores from all sims to date including breakdown of energies (as per the previously assigned tasks) ASAP as priorities.
RSK: I will get this done fast
RSK: I tried creating 2D Ligand interaction diagrams using the tool available in Schrodinger. However, it sounds that this tool has a limitation in handling multiple ligand at the same time ( Ie you can define either NAD+ or the peptide as the Ligand). Hence, I am currently unable to annotate the C-N ( ribose ) and Acetyl Oxygen distance. I have attached a sample plot which I prepared for Sirt3/NAD+/AC-ACS 4FVT loop vs 4BVG loop.
Avg_Ligand Interaction.pdf
RSK: There is an workaround for this. I can request for a free version of LIGPLOT+ software for PMC-AT which is available from EBI (http://www.ebi.ac.uk/thornton-srv/software/LigPlus/) and then we can fix this issue . The earlier protein-ligand 2D interaction plot which I prepared for the 4FVT and 4BVG crystal structures were taken from the PDBsum database which uses the LIGPLOT+ software for generating the images. PDBsum database has protein-ligand interaction diagram of only complex structures deposited in the PDB
RC: Ok that's fine, we can try that. For now in the existing ligand interaction plots just write down the average distance.
One other issue with these diagrams is that they don't clearly indicate the residue numbers for some of the side chains depicted.
In addition, some of the residues I was curious about are not appearing in the diagram. For example, Phe157.
RSK: I need to check closely about Phe 157. The cut-off distance used here for computing the non-bonded interaction was 4A may be we can extend it to 5A to show other important residues
Also, please provide the corresponding diagram for the INT:NAM complex (is the INT complex diagram already available in PLOS?)
Perhaps we will see Phe157 in the latter diagram. Note that it seems to contact NAM in the 3d MD average structure
for INT/NAM that we have reported in the draft paper. On the other hand, it was also not visible in the PLOS NAM ligand interaction diagram.
In addition, a loop Arg (Arg34 in Sir2 numbering) has been described in some literature as interacting with the ligand upon loop closure.
Are the above diagrams from the full simulation or the 1 ns simulation (i.e., are they corresponding to the same structures you sent me on Fri or not?)
RSK: The above diagrams are from the Average structure generated from the 12 ns run
I plan to discuss issues with the results with you shortly, depending on when some of the necessary items above are complete (perhaps tomorrow). Please let me know when you anticipate sending the above.
RSK: I can get you all these results by Wednesday.
RC: Ok, as you send the updates in parts I will determine when I have sufficient info to have a meaningful discussion and then set the time.
RC: Any updates available on this? Please let me know what you have available now and what remains. If you have some items complete now you can send them over so I can start review,
but I don't want to slow you down in case none are finished.
According to schedule it appeared you would be in the process of scoring the long trajectories now but the various ligand interaction diagrams would be largely finished, and some of the
recently indicated simple calculations including RMSD calculations/associated structure alignments of ADPR and NAM moieties for ternary and intermediate complexes, breakdown of energies, pending prime scoring of ternary closed loop structures, etc would be finishing up as well.
RSK: Amber MM/PBSA and MM/GBSA rescoring for all the trajectories are complete
a) Sirt3/Product complex ( 4FVT)
b) Sirt3/product complex (4BVG loop)
c)Sirt3/Ternary complex (4BVG loop)
I can turn in the prime energies for INT/NAM starting complex today.
RC: Ok, I think you mean you will be uploading a-c) today.
How about other ligand interaction diagrams including INT, and other recently indicated simple calculations mentioned above?
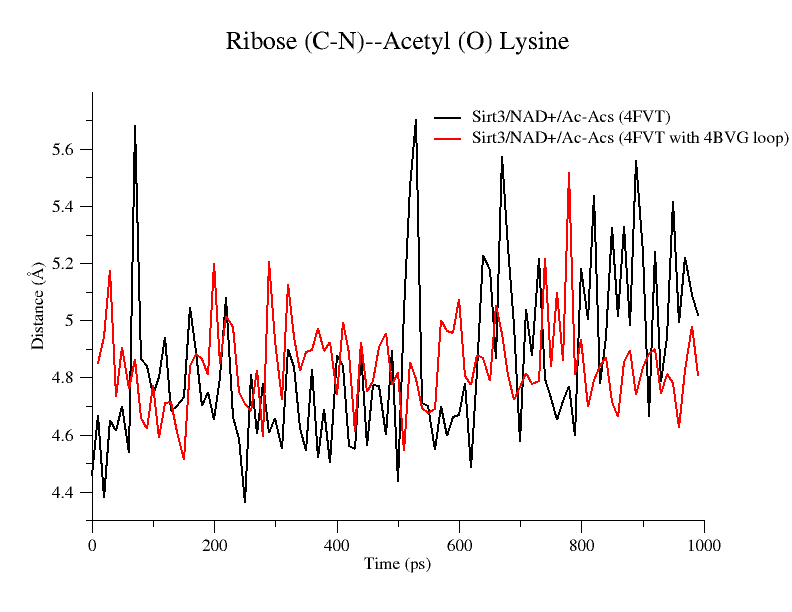
RSK: I have plotted the distance between the ribose C-N carbon (anomeric carbon) and acetyl oxygen using the 1 ns trajectory data for Sirt3/NAD+/Ac-CS2 with a ternary loop (4FVT) and with a "intermediate loop”. The data doesn't show any difference in their mean distance (4.84 Amg. Although it’s too short a time scale to meaningfully conclude). However, a close analysis of the 1ns trajectory reveled the ribose plane to be rotated towards the Acetyl group of Lysine in Sirt3/NAD+/Ac-CS2 and the NAM part of the NAD+ is in plane with the ribose in the ternaray but "out of plane" in the Intermediate loop model. I have attached images to illustrate it.
I believe that your idea here was to see if the ribosyl ring in NAD+ gets reoriented so that it is poised for the deaceylation reaction that involves the breaking up of the glycosidic bond between nicotinamide and the N-ribose of NAD+, which subsequently leads to the formation of a covalent "SIRT3-alkylamidate intermediate".
If that was the intended reason the calculating the dihedral angle ( order parameter) between the Acetly lysine oxygen , the anaomeric carbon atom of the Ribose and the carbon that links the phosphate oxygen and the ribose ring could be a more reasonable order parameter to capture the change that the orientation of the ribose ring in the presence of an intermediate loop conformation. I see that these three atoms are almost co-planar in the "intermediate complex" evident from 4BVG Xtal structure Please advise me if we should go ahead with current distance based order parameter or should we consider a more refined order parameter that could capture the reorientation of the ribose group.
RSK: The distance plot and the images are attached. I will annotate the average distance (4.84 for both systems) on the Ligand interaction diagram as suggested by you.
angles_distance.pdf
RSK: Revised schedule
revised_schedule_V8.docx
RC:
-Regarding interatomic distance, I was suggesting that the be annotated in the ligand interaction diagrams for MD average structures (misc task 1) and
thought those tasks might be done together early on.
You can prepare a distance plot if you like, but it should still be annotated as such in the ligand interaction diagram as well.
-According to the schedule it appears the ternary closed loop full simulation is to be finished by Mon?
-Will advise on Jul 13-14 tasks
RSK: The 12 ns simulations for a) 4FVT with coproduct/peptide and b) 4FVT with 4BVG loop and coproduct/peptide are complete.
I expect the 12 ns simulation for the ternary closed complex ( Sirt3/NAD+/Ac-CS2/ with 4BVG loop ) to be complete by Monday as per schedule. I will launch the job before I leave for the day today.
Currently only this simulation will be running, but I will be running some MM/PBSA and MM/GBSA CPU calculations tomorrow for the above mentioned systems A and B ( as per the schedule).
I hope that will not cut much in to the estimated speed of 3.5 ns/day/system.
RSK: I have updated the Amber MM/GBSA and MM/PBSA results from the 1ns simulation for Sirt3(4FVT)/NAD+/4AC-CS2/4BVG loop on the xls sheet. I am attaching it for your perusal.
MMGBSA_scores_V6.xlsx
RSK: Break up energies of the Amber MM/GBSA and MM/PBSA energies
MMGBSA_breakdown.xlsx
RSK: Simulation Status
I was supposed to have completed a 1 ns simulation for 4FVT/ternary complex/4BVG loop by July 1st as per the schedule. However, I ran in to many issues while setting up MD simulation (In particular reproducing the exact parameters for NAD+ and the Ac-Lysine of the peptide proved to be a challenge). Finally, I located the parameters used by Ping for NAD+ and Ac-Lys of the peptide. The simulation has been submitted today (7/6/2016) morning. I am now confident that we can compare the 4FVT ternary complex simulation (PLOS paper ) data with the current simulation data. I made sure that partial charges, force field and nonstandard residue parameters employed here are the same as in PLOS paper data as we intended to compare it as a pair ( ternary complex closed vs open loop).
Once I get these values I will upload the revised schedule along with the data for Miscellaneous Task 1.
I expect the 12 ns MD simulation for the below mentioned pairs to be complete by tomorrow.
a) 4FVT with coproduct/peptide
b) 4FVT with 4BVG loop and coproduct/peptide
Once this gets complete. I will launch the extend 12 ns simulation for 4FVT with 4BVG loop ternary complex on Thursday.
RC: Previously you mentioned that PL had parametrized aadpr and run md simulations previously for coproduct complex. I believe you mentioned you reran these but used the same parameters and charges?
Why did the ternary complex pose such a challenge with respect to nad and ac peptide parameters? Was it the only case where you needed to replicate PLs parameters? See my earlier questions regarding statement in Plos that parameters for NAD developed by walker and pavelites were adapted and parameters for Ac-Lys developed by Papomokos were adapted.
When you get a chance, please summarize any differences between the parametrization approaches used for these vs the other ligands.
RSK. In fact the references which you pointed out helped me in locating the libraries which Ping has used. In fact I paratmaterized AADPR using the same parameters form Pings data. However, NAD+ and Ac-Lysine were not done before. I wanted to be extra careful here because we need to compare the numbers between closed vs open. ( PLOS data vs current data). Sure, I will update it on the wiki when i find time.
Please confirm following:
-1 ns closed loop ternary and updated schedule including the misc tasks and scoring (on CPUs) to be provided by Th
RSK: Updated schedule complete. 1 ns job submitted today. I expect it to be complete by Friday ( 3 Jobs are currently running on the GPU )
a) 4FVT with coproduct/peptide (11 ns job)
b) 4FVT with 4BVG loop and coproduct/peptide (11 ns job)
c) 4FVT with 4BVG loop ternary complex ( 1 ns job)
-Both 12 ns coproduct simulations will complete by Th. Scoring to be set up thereafter.
RSK: I am expecting it to be complete by tonight ( I checked this morning and I see that 9.5 ns to be complete of the total 11 ns run. ( Please be noted that we are running multiple jobs (3 jobs) here and this has cut into the estimated speed of our earlier bench mark performance that 3.5 ns/day for a single system. In case if you feel that some systems should be given priority over other , please let me know, I can kill an less priority job and restart that simulation once our HIGH priority jobs gets completed)
-Then 12 ns ternary closed loop simulation to be started on Th. Will scoring for 1 ns be completed before this, by Th?
RSK: I expect to set up the two MM/GBSA scoring on Saturday so that we can have the data by Monday. I have listed MM/GBSA run for 4FVT/Ac-peptide/NAD+ with 4BVG loop 1 ns simulation for Friday along with other items Miscellaneous tasks .
RSK: I have copied below the questions you had send out to me via email on Sunday (7/3/2016) . My answers to them are also embedded within. This is to ensure that its is document on the wiki space for convenience. As per your comments I will revise revise the work schedule and upload it by the end of the day.
RC:I am seeing several issues with the revised schedule that need to be addressed immediately.
1) I thought I had mentioned that the 2-3 12 ns simulations we would be proceeding with would be:
a) 4FVT with coproduct/peptide
b) 4FVT with 4BVG loop and coproduct/peptide
c) 4FVT with 4BVG loop ternary complex
4BVH were not the preferred simulations. I believe I mentioned this. Please see:
"-After receiving the above results we will confirm that the pair of 4FVT based coproduct complexes should be subjected to 12 ns simulation (this will most likely be the case). "
"We will probably need to run 2-3 12 ns simulations (open, closed coproduct and closed ternary) thereafter so there will be time for other tasks while those run."
All of these (open, closed coproduct, and closed ternary) use 4FVT protein structure.
RSK: I think I inadvertently messed up the systems while editing my schedule .Sorry about that. In fact system A and B which you did mention in your email on Saturday had been subjected to 12 ns simulation on Saturday morning. They are currently running and I am checking them intermittently. I expect the 12 ns MD runs for systems A and B (4FVT with coproduct/peptide and 4FVT with coproduct/peptide) to be complete by Tuesday night (~ 3.6 ns per day is what I am able to achieve when I run two systems in parallel). I did realize that 4BVH was of less priority.
RSK:Once this MD job gets completed (2 systems I am running in parallel) I will launch the “4FVT with 4BVG loop ternary complex” simulation. I will revise the schedule according shortly.
RC:The point of doing the 1 ns analysis was to avoid the need of doing the long simulations for complexes that we would not present in the paper.
Not sure why it was indicated on wiki that 4BVH simulations would proceed from Jul 1 evening.
I would like to know when a,b) will be started and how long they will take.
RSK: System A and B (4FVT with coproduct/peptide and 4FVT with 4BVG loop and coproduct/peptide) will be over by Tuesday night. I expect to provide you the MM/GBSA and MM/PBSA results by Thursday (7/12/2016). A single MM/GBSA and MM/PBSA re-run would take about ~ 4-5 hrs for a 12 ns trajectory.
RC: I did not comment further on 1) on Fri since you had not yet finished the ternary complex 4FVT with 4BVG loop 1 ns simulation.
Please update with the results from this.
RSK: I realize that I was supposed to get this task completed by Friday. However, I faced many issues here particularly with parameterizing the NAD+ and the Acetylated Lysine of the peptide. Please be noted here that the bottle neck here was that I need to make sure that the parameters which I am using here are the same as those used by Ping used for the Sirt3/NAD+/Ac-CS2 peptide with 4FVT (PLOS paper). I am not sure how he parametrized those nonstandard residues (I have mentioned this issue earlier that Pings documentation lacks clarity about the treatment of non-standard residues), so I had to manually check that topology and parameters file to confirm if the parameters are the same. Only if the parameters are same we can compare the numbers between closed and open ternary product complex. I will update the results for this shortly.
RC: I did not see the miscellaneous tasks under misc task #1 addressed in the short-term schedule. It was stated that these could be done immediately, possibly starting Fri/this weekend since the data needed for them was mostly available. (I mentioned this couple of times over the last few days including Fri. Please see my last email.)
RSK:I think that you are talking about the miscellaneous task (less priority) “Related structure alignment task: align the PLOS INT/NAM MD average with that from the latest INT/NAM simulation (closed loop), check RMSDs, including that of NAM, acetyl-Lys and rest of ADPR. Note energies cannot be compared since PLOS used 4BVG.”
I am bit confused here, could you please elaborate the miscellaneous task 1 which I have copied above. I believe that PLOS paper deals with SIRT3:Ac-CS2:NAD+ (4FVT) MD simulation . Ping has done simulations using Sirt3: Int:NAM starting with 4FVT and 4BVG loop. I calculated and reported the RMSD (open vs closed) for the loop region, global RMSD, ADPR moiety, peptide part earlier for these simulations. I didn’t carry out any INT/NAM (Intermediate product) simulation. However, I did calculate and report the MM/GBSA and MM/PBSA energies for time t=0 to t=1 ns from Pings previous trajectory.
RC: I am referring to the following comment in the document revised_schedule_v6_RC_6-29.doc below: "For the ternary complex open loop (4FVT) and closed loop (4BVG) MD averages, annotate the interatomic distances between ribose C-N carbon and acetyl oxygen. Do this for the ternary complex open loop MD average first since we already have the MD average for the full production simulation. Then report it for the 1 ns simulation MD averages for open and closed loops. Finally, report it for the closed loop MD average from that full production simulation."
RSK: Okay, got it. Thanks for clarifying.
RC: I had mentioned recently that I am interested in more details of the full production run scores for ternary complex with open loop (from PLOS). I would like to know whether you have access to the scores for complex, receptor, ligand (not just binding energy) from the simulations reported in the PLOS paper, specifically the 4FVT open loop ternary complex simulation. I noted they are not reported in the paper itself.
If so, please report them on wiki.
RSK: You are right. These score are not reported in the paper. However, I got the binding energy values from the paper and reported it on the wiki. I see that the MD trajectories for these systems are lying there. I can look into these directories and see if the scores are there. If not, I can get these numbers by rerunning the trajectory.
RC:Related, I noticed you reported data for both NAD+ and peptide ligand for the ternary open loop 1 ns simulations. If you have the complex, receptor, ligand scores for both of those please report as well, esp if you think it will take time to retrieve the data for the full production run. If you have these data handy for the other complexes you are studying as well please let me know. I may then ask you to report some of those too. (Here, I am particularly interested in the INT/NAM simulation with the two different ligands. Again the 1 ns results would be sufficient for now.)
RSK:Yes, I do have the values for the Ligand, protein and complex from the Amber MM/GBSA and MM/PBSA scores.
RC: Please note that this task is of lower priority than the others listed above and on the schedule.
RSK:Okay, got it. Will list it under low priority task.
RC: Regarding the MD average structure scoring, you mentioned both before and now that it would take 2-3 hours per complex to rescore.
RC: I took that to mean whether it was the final frame or the MD average. Is that right?
RSK: It was the final frame. So the necessity to construct an average structure and energy minimize the average structure didn’t arise. The average structure will have usual bond length, angles and torsion because it is simply an average of XYZ coordinates of multiple frames. Hence, it needs to be processed before any calculations can be undertaken on the average structures. That’s the reason I hinted that it may take more time that final snap short scoring. Also, Ping has constructed averages only using the last 10 ps of the trajectory (essentially it is an average over 10 frames)
RC: It appears you already rescored all the final frames? If so please remove the MD average structure scoring from the task list tentatively, or put it as an optional task at the very end.
(Regarding 10 ps averages, there may be problems forming MD average structures from long trajectories. I will consider whether we need to form and score MD average structures and revert in the event that we do. The task was proposed because rescoring the whole trajectory with opls/vsgb was considered too time-consuming.)
RSK: Yes, I had scored all the final snapshots.
RSK: Yes, you are right here. The last 10 ps many not be an ideal representative snapshot of a time averaged structure.
RC: Note also that in revised schedule replace the md average structure prime rescorings with the first few misc tasks
I prioritized some misc tasks in my latest schedule update. I know you did not intend these to be done in order. However please observe the changes I made to the order of the first few misc tasks and make these the priority misc tasks in your schedule
RSK: Okay, once the miscellaneous task 1 gets clarified I will schedule it.
RC:You mentioned 2 gpus for MD simulations. What about other tasks like rescoring? Are you running those on cpus?
RSK: Yes, I plan to use the CPU on the pmcat-gpu node. I believe that using the CPS will not cut in to the GPU speed. pmcat-gpu1 is the only workhorse for computing as of now. I don’t see that Amber can be invoked form the other nodes at least slave003 which I did check.
RSK: Updated work schedule.
revised_schedule_V7.docx
RSK: Updated xls sheet with scores ( you can ignore the xls sheet which I uploaded yesterday ).
RSK: A quick update of my work status.
The only system for which I need to compute the MM/GBSA and MM/PBSA score is Sirt3/NAD+/Ac-peptide/ with 4BVG loop. The modelling of the system is complete (grafting, side chain repacking and prime minimization). I will send out the results for the 1 ns simulation tomorrow.
I believe that we will have the scores for all systems by tomorrow.
MMGBSA_scores_V5.xlsx
RSK: Updated xls sheet with scores. I expect this sheet to be completely filled up before Friday.
I ran in to issues when I scored Sirt3/INT/NAM intermediate complex using Prime. It took me time to figure out the issue and fix it.
The issue was Prime was not able to handle the INT (Intermediate ) because the Intermediate was modelled by Ping by introducing a covalent bond between OADPR and peptide. Unfortunately, Prime kept parsing the OADP atoms as small molecule and the peptide part of INT as a protein (In other words it didn't treat INT as a single molecule). I had to then manually edit the PDB file and had to rename all the peptide amino acids as OAD ( small molecule ) and forced it as to be treated as "HETATOM" and not as "ATOM" record in the PDB file. Then the whole INT was parsed by Prime as small molecule.
MMGBSA_1ns_eq_6_22_2016_V4.xls.xlsx
RSK: Revised xls sheet with Prime energies.
MMGBSA_1ns_eq_6_22_2016_V3.xls.xlsx
RC: Please add rows for ternary complex (NAD+and peptide ligands), to be filled in with data as they become available.
We can also include rows on full production runs in this xls as they become available.
Apparently it should be possible to fill in ternary open loop full production run row at this time (including both complex and binding energies) given data from PLOS paper; 1 ns may require rescoring.
RSK: I will turn in the 1ns re-scoring results by today. Yes the fill production value can be adapted form the PLOS paper.
Priority is to set up the closed loop ternary simulation (4FVT with 4BVG loop). Please update regarding status of this shortly and whether you are encountering any of the same issues mentioned by PL in his brief report mentioned below.
RSK: Apparently it looks like I will have to model the system form the starch. Anyways, i will try to set it up today ( if any issue arises I will get it done over the weekend and get the results by Monday).
Run this simulation at least over weekend (if not complete today), and provide the single point prime energies using MD average structures (see comments below) as time permits over next couple of days, given that you mentioned these would take 2-3 hours per complex.
We will probably need to run 2-3 12 ns simulations (open, closed coproduct and closed ternary) thereafter so there will be time for other tasks while those run.
RC: Switch INT and NAM simply meant the numbers were not in the right rows as far as could tell (NAM number was provided for INT and vice versa). By row order I meant simply that for FVT based structures, open loop was provided first, whereas for BVH based structures, closed loop was provided first.
RSK: For some weird reason dont see the above comment getting listed on the wiki in the Edit mode. So i copy/pasted it here so that I can provide answers.
RSK: I believe the numbers for INT and NAM were placed in the right row.
RSK(6/22/2016); I have updated the xls sheet with the MM/GBSA and MM/PBSA score for both equlibration and 1 ns short simulation for the following systems
a) Sirt3/INT/NAM - 4FVT recp with native loop ( MM/GBSA and MMPBSA score provided for INT and NAM)
b) Sirt3/INT/NAM - 4FVT recp with 4BVG loop ( MM/GBSA and MMPBSA score provided for INT and NAM)
c) Sirt3/2’-OAADPr- 4BVH rec/ loop
d) Sirt3/2’-OAADPr- 4BVH recp/3GLS loop
e) Sirt3/2’-OAADPr/Ac-cs2 deac-4FVT rec/loop
f) Sirt3/2’-OAADPr/ Ac-cs2 deac -4FVT rec/4BVG loop
System G and H , need to be done next.
G)4FVT with NAD+/peptide -- Native loop ( Data aldready avalaible PLOS paper)
H) 4FVT with NAD+/peptide loop from 4BVG ( Data availability not sure ??)
Moving on to the next two items.
MMGBSA_1ns_eq_6_22_2016.xls.xlsx
RC: I think your NAM and INT rows need to be switched.
Please present open/closed data in same row order for BVH and FVT-prepared simulations for clarity.
I am planning to ask you to rescore a single frame from the 1 ns simulation from
each simulation using VSGB (will advise on details shortly). Please let me know roughly how long this will take.
Please update me once you know about data availability.
RSK: The revised sheet is attached.I guess I understood your "same row order " and "switch INT and NAM" instruction correctly. If not please let me know I will fix it.
I guess each MM/GBSA calculation for a single frame should roughly take 2 hrs. Steps include
a) Convert the NAMD dcd file to a pdb file
b) process the PDB file using the "protein preparation wizard of Schrodinger". It includes OPLS 3 based minimization of teh complex.
c) Split the PDB file in to LIGAND and RECP
d) save the ligand in to a maegz file and the receptor in to a PDB file.
e) Run MM/GBSA in prime
MMGBSA_1ns_eq_6_22_2016_V1.xlsx
RC:
- After clarifying data availability/setting up G,H) as needed, please provide complex energy from full production run for ternary complex with open loop (G), which we should already have.
RSK: I will be able to provide you an answer for this by today evening .Prima facie it looks like that we may not have data for 4FVT with NAD+/peptide loop from 4BVG (system H).
Although Pings directory is named as " MD_4FVT_with_4BVG_loop" but visualizing teh trajectory it sounds to be an Sirt3(4FVT/INT/NAM) with 4BVG loop. I will check other trajectories also, since you told me that you rember Ping started the simualtion.
RC: When I indicated Ping started work on this I was referring to the document "SIRT3-NADp-ac-LYS complex and loop substitution". He may not have started the simulation it seems.
Note you can provide the complex energy for G above from full production run (I'm asking for this since earlier your provided binding energy from plos on wiki below, but not complex energy -- it may be reported in plos).
- Then please provide VSGB scores for each starting structure used for MD (for all complexes in the spreadsheet), which I assume were minimized using VSGB.
Yes, I do have the score for all the complexes ( Prime score uses OPLS 2005/ VSGB) . I will send it out today in a updated xls sheet
RC: Yes, I mean OPLS/VSGB, whatever name they are using for the composite MM-GBSA scoring function used for scoring these days.
This should not take long. Please confirm that you already have these scores, at least for complex. Provide complex and binding energy scores in spreadsheet and indicate what is ligand in each case.
I don't have the binding energy score . the binding energy score is the Prime MM/GBSA score.But, I can get it ( should it schedule it to my task list ?)
RC: Yes we'll need that for comparison to Amber.
-Then I may ask you to also rescore one structure from production runs of each complex with VSGB. Here we may choose either last frame or MD average as we are only scoring one structure in each case. Let me know if the MD average structures that you have reported can readily be used for VSGB calculations or if additional time consuming steps would be required to prepare the files. In the latter case you should rescore the final frame from each 1 ns production run for now.
Please provide an idea of how long this will take.
RSK: A single structure should be some where between 2-3 hrs.
-After receiving the above results we will confirm that the pair of 4FVT based coproduct complexes should be subjected to 12 ns simulation (this will most likely be the case). This should ideally run over the weekend. If there an issue of CPU/GPU resource allocation you should first complete the equilibration and 1 ns scoring for the ternary complexes above
.RSK: I have a question regarding resources. I have been always using the pmc-at gpu node. Can i use the other nodes ion the cluster ( I belie that are MPI enabled). I have not checked them. I chose pmcatgpu1 node blindly because I see that Ping had also used the same one. In that case we can schedule multiple MD jobs. Rescoring can be ideally done on CPU and MD simulations on GPUs to save time.
RC: Agreed (Ping and I had discussed this, but I believe he never really used the other nodes), but don't spend more time working out networking issues right now.
We will deal with the pbs server after finishing work on this paper, not needed right now.
The immediate rescoring are just single structures, so I assume that is mostly manual prep time.
When you get to the point where you are looking at using the other nodes, please provide a status update on them, but don't delay the other immediate tasks to start this.
-Then we will also need to run the full 12 ns simulation on the ternary complex with 4BVG loop (after structure preparation, equilibration, 1 ns production are complete).
RSK:Okay
RSK: Revised schedule updated based on your comments from version 3 and 4 and incorporating the ternary complex simulation pair ( 4FVT and 4FVT/4BVG loop)
revised_schedule_V6.docx
RC: Schedule with minor revisions to miscellaneous task order below, as indicated.
As discussed on email pls update rest of schedule shortly so we can get an idea of the extent to which work
has been pushed back due to need to run the 2-3 long MD simulations in series (assuming that is the plan) and whether
we can make use of that time to finish other tasks on the schedule.
revised_schedule_V6_RC_6-29.docx
RC (6-29): Schedule above has been further edited slightly. See #1 on misc task list since it includes some items directly related to current work.
Please provide the updated xls including the full production complex energies for ternary open loop, 1 ns energies for ternary open loop.
See also my comments made earlier regarding prime rescoring of MD average, which should be scheduled if not already done.
RSK: Updated results from MM/PBSA and MM/GBSA calculations. I will keep updating the list as the MD jobs and the MMPBSA run gets completed.
MMGBSA_1ns_eq.xls.xlsx
RC: Please report binding energies for INT rather than NAM in the INT:NAM results. Please indicate when pending data listed in xls are expected.
RSK: The pending data on the xls will be updated by tomorrow afternoon. I have setup the system already. The equalization and simulation will run overnight and I will get the values by tomorrow afternoon.
RC: Ok I look forward to receiving those. I will have some additional follow up work and comments/queries thereafter based on results.
I assume you will also be reporting binding energies for INT rather than NAM as indicated above.
RSK: MM/PBSA and MM/GBSA results of the equlibration phase and a short 1ns MD simulation for SIRT3/2'-OAADPr/Closed loop (4BVH -native) and SIRT3/2'-OAADPr/Open loop (4BVH and loop grafted from 3GLS apo enzyme).
MMGBSA_1ns_Eq_Data_4BVH.xlsx
RSK: A revised schedule which incorporates the current status of the work and a estimate of the time frame for completion.
revised_schedule_V5.docx
RC:
-- I don't see the structure prep, simulation and scoring tasks for new simulation "b" below:
"b) Setting up closed loop simulation (equilibration phase to start) for 4FVT with NAD+/peptide, following side chain optimization, then running scoring
(miscellaneous tasks can proceed during this simulation). Report equilibration phase scoring first."
Please add them to the schedule as per my comments on this task.
-- Regarding miscellaneous tasks, I don't think you accounted for all the comments made regarding misc task schedule
in my latest marked up task list sent last week.
The misc task schedule/order needs to be edited per those comments.
Please make these revisions (you can make them after reporting the latest batch of scoring results or after you start work on simulation b) below.
RSK: Okay, i got it. I thought that 4FVT with products (2'-OAADPR and Deac Ac-Acs peptide ) with its native loop ( ternary loop ) and 4BVG loop (intermediate loop are the ones we are interested ). Sorry, I overlooked the
system 4FVT with NAD+/Ac-Acs peptide and 4FVT with NAD+/Ac-Acs peptide with 4BVG loop.
Will revise the schedule once I send out the MM/GBSA and MM/PBSA results for the first batch.
RC: Note that the results for b) will be compared to the corresponding scores for a) below. If you need to do analogous scoring
for a) (scoring for the full sim was reported in PLOS) you should schedule that as well. But the results for b) should be reported as soon as they are available.
RSK: Apparently, it looks like Ping has carried out both the simulations. I see that there are two directories "MD_4FVT_tern" and "MD_4FVT_with_4BVG_loop". I guess that MD_4FVT" will have all the data reported in the PLOS paper and the MD_4FVT_with_4BVG_loop" contain the data for the model which we intend to model.
In that case I guess that Sirt3/NAD+/Ac-ACS peptide with native and grafted 4BVG loop task would be easy.
Anyways, let me analyze the trajectory more closely before I come out with a strong statement. In any case I will have to do analogous MM/GBSA and MM/PBSA scoring on the trajectories even if the trajectories are the same as we intended to model.
RC: He started to do it but as I may have mentioned earlier, his report in dropbox was cut short and he indicated the energies of open/closed loops were not comparable. Yes,
you should look into his data and report once you get to this point.
RSK: MM/GBSA and MM/PBSA results from Open vs Closed loop conformation ( Sirt3/2'-OAADPr/ native loop -4BVH and Sirt3/2'-OAADPr/ closed loop grafted form 3GLS ) for the product complex shows that the binding affinity of the procuct (2'-OAADPr) is enhanced for the native CLOSED loop over modeled open loop conformation .
Please note that these are preliminary findings based on an Equlibration simulation. However, I would say that the story may not change even if we run longer simulation because the binding energy difference is marked here.
The second caveat we need to bear in mind is that the product (2'-OAADPr ) was not docked to the modelled open conformation, instead we faithfully copied the coordinates of 2,-OAADpr from its native complex(4BVH ) on to the modeled open loop complex ( generated using apo 3GLS) assuming a similar binding orientation. This protocol is consistent with the 4FVT and 4FVT/4BVG intermediate models created earlier.
I mean to convey here that the short simulation may not have adequately sampled the internal degrees of freedom of the PRODUCT/LIGAND here. It may be have been trapped to the initial conformation of the ligand/product.
Open_closed_product complex.xlsx
RSK: I have attached the revised schedule for the paper based on your feedback.
The revised schedule accounts for the modelling of various complex ( 4FVT- with AADPr /peptide product and 4FVT-with 4BVH loop and AADPr /peptide product, selection of pair of systems for MD simulation based on initial MM/GBSA scores and a miscellaneous task that includes analysis and presentation of the side chain validation data generated by Ping.
revised_schedule_V4.docx
RC:
Before starting miscellaneous tasks, we need to do two additional tasks:
a) MM-GBSA scoring for 4FVT with NAD+/peptide, open loop (ternary complex; trajectory available)
RSK: I think that the said complex has been subjected to MD simulation by Ping and the values are reported in the PLOS paper.
MM/GBSA for Sirt3/Ac-Cs2/NAD+ -65.63 kcal/mol
MM/PBSA for Sirt3/Ac-Cs2/NAD+ -5.35 kcal/mol
RSK: I am not sure here, when you say "OPEN LOOP" do you want to graft the Intermediate loop conformation form 4BVG onto this ternary complex and proceed like we did for the other systems ??? Just wanted to make sure if we are on the same page regarding the "open/ closed loop semantics here.
RC: Are the above numbers for NAD+ ligand binding energy? Was peptide ligand scored as well?
RSK: the above numbers are for NAD+ in Sort3/Ac-Cs2/NAD+ complex
RSK: I guess that Sirt3/Ac-Cs2 were treated as receptor and the NAD+ was treated as the Ligand here. The affinity of the peptide was not scored as evident form the PLOS paper.
RC (6/16): Ok, we can always score peptide if needed.
RSK: Yes, thats right provided we have the trajectory details for the old paper. I believe that it should be in GPU node.
Open loop means the native 4FVT loop. Closed means 4BVG loop.
b) Setting up closed loop simulation (equilibration phase to start) for 4FVT with NAD+/peptide, following side chain optimization, then running scoring
(miscellaneous tasks can proceed during this simulation). Report equilibration phase scoring first.
RSK: Okay, as planed and listed on my schedule I will report the MM/GBSA scores first after completion of the equlibration phase, to decide on the system for production simulation.
RC: I also mean to report equilibration phase scoring for the new simulation b) before moving on to production simulation for that.
RSK Thats right, we have to model two systems with
4FVT native with AADPr and peptide - Limited modelling ---- Task completed
4FVT grafted with 4BVG loop complexed to AADPr and peptide - Task partially completed : Will be completed by today ( NB: side chain repacking ( ~ 7.5A radius ) along with prime minimization of the repacked side chains takes ~ 2hrs
RSK: Setting up equlibration MD for the purpose of a short MM/GBSA scoring to decide on which system to consider for simulation
We have to consider THREE systems
i) 4BVH with 3GLS grafted loop : STATUS - MD setup completed, have to test if the set up fine ( run a minimization and check the energies and see if the system is stable ie, it dosent explode during the
equilibration
ii) 4FVT native with AADPr and deac-peptide - STATUS - working on it - will be complete by today
iii) 4FVT with loop grafted form 4BVG complexed to deac-peptide - STATUS: will be completed tomorrow morning
RC: Based on schedule, I assume you will be reporting scores (complex, receptor, binding energies for one or two ligands) from at least i,ii) today, with iii) following shortly after.
I would need some time to review these in order to settle on the production simulations etc.
RSK:I can provide you the numbers for system 1 today ( equlibration result). This task is completed
Also, I will try my best to turn in the results for system II alt least later tonight. ( I am setting up the system now.)
(a) will be used to compare with INT/NAM open loop, in particular with respect to binding energies of ligands, given that open loop is expected to be less stable than closed for INT/NAM but not for NAD+.
(Note that the scoring may already be available; it was reported in PLOS for one simulation, though there may have been slight differences in structure preparation (check this and report).
RSK: Yes the MM/GBSA and MM/PBSA scores are reported in the paper for Sirt3/Ac-Cs2/NAD+ ternary complex( I guess it is 4FVT with native loop -Ternary loop ). The values are reported above
RSK: I believe that you want some thing similar to that ie, (Sirt3/Ac-cs2/NAD+ grafted with a intermediate loop (4BVG)
RC: Yes
(b) will be used to verify that the closed loop is less stable for ternary complex and hence consistent with structural data.
(Note that PL was apparently setting up this simulation when he left; see the latest note in his dropbox where he indicated a possible problem with structure preparation.)
RSK: Okay, I believe you are taking about the Sirt3/Ac-cs2/NAD+ grafted with a intermediate loop (4BVG). I will look in to it.
RC: Yes
The reason consistency is important to check asap is that there is some evidence from the INT/NAM simulations that there may be considerable energy errors in scoring/ranking loops that
may lead to qualitative discrepancies with experimental data. For example, for the apoenzyme, open loop is apparently more stable, but at least in the presence of INT/NAM
we see that the closed loop is predicted to be more stable to an extent that cannot be explained on basis of interactions with ligands alone. Of course, for INT/NAM open loop simulation, we observe
global changes in the protein structure that are not observed for the closed loop simulation, and we did not run a separate trajectory for receptor alone. Hence it may not be appropriate to draw conclusions regarding energy errors or accuracy of loop scoring based on the INT/NAM pair of simulations alone.
We need to explore this issue of consistency further before deciding what to present and how. Depending on what we find, we might focus on energy comparisons between different complexes having the same loop conformation, or between binding energies
for different loop conformations. This also makes it more important to consider use of the 4FVT-based structures for all calculations including those for coproduct.
RSK: Okay I got your point.
RSK: We are following the same protocol implemented in the PLOS paper. So consistency is there.
a,b) have been mentioned in attached schedule doc below. Please indicate how long they would take so we can make a judgment on how to proceed
given the results. While you do these, I can review the recent equilibration results and decide on next simulation.
RSK: The new task which you has listed now Sirt3/Ac-Cs2 peptide/NAD+ with a open loop???? ( Need to conform if you are indicating 4BVG - Intermediate loop or 3GLS- apo enzyme open loop here???) would take a complete 2-3 days of work for modelling the system (grafting loop/side chain prediction/energy refinement of the modeled side chain followed by complete prime minimization and MD system set up for equilibration
RC: You said a) above is done, hence b) remains and that is 4FVT with closed (4BVG) loop
In addition, let me know how difficult the following would be:
c) rescoring single structure (e.g., frames to be specified) with VSGB/OPLS ff,
for each of the complexes studied
d) rescoring binding energies for each of above
RSK: Re-scoring a single frame in Prime using the VSGB/OPLS2005 ff would be ~ 30-45 min task. But re-scoring all MD frames will a more time consuming , because we need to writes scripts for that and automate it.
RSK: Piratically prime MM/GBSA can be used only a single frame "rescroring " method and cannot be run on multiple frames from a trajectory , because each frame form the trajectory (dcd frame has to be converted in to PDB file which in turn needs to be converted in to a mastero compatible mae/ maegz file and the subjected to protein prep and then finally run MM/GBSA which involves a simple first order minimization. Although theoretically possible, I would say it is exhaustive.
RC: That's what I thought, hence we would only score a single frame in each case first, after which we will determine whether anything else is needed.
These are meant to explore robustness of results with respect to choice of energy function (and determination of whether apparent energy errors can be corrected with VSGB).
They might be done after analyzing the results from a,b) as needed. Note that VSGB has been developing in the context of applications like loop prediction so we want to see if it can
provide more consistent results, to start for single point calculations.
RSK: yes, you are right VGSB has been developed in the context of loop prediction ( particularity for long loops). But, recently Schrodinger has been promoting Prime MM/GBSA ( ie, OPLS ff with VSGB continuum solvent model) as a single snapshot re-scoring function for protein-small molecule complex and for computational Alanine scanning .
In case there are irreconcilable energy errors with loops, the side chain prediction validation work will at least show few energy errors for fixed backbone conformations.
Note for certain smaller segments of these same loops, we saw reasonable predictions in past work.
revised_schedule_V4_RC_comments.docx
RSK: Will revise the schedule on your comments and upload it tomorrow. I see that the new system ( Sirt3/Ac-Cs2-NAD+) needs to be added to the schedule now. One system Sirt3/Ac-Cs2/NAD+ with ternary loop (4FVT) has been done earlier, by ping but I will had to carry out one more simulation for completeness. I guess it is one modelling and simulation.
RC: Yes, if a) is already done, only b) remains.
RC (6/9): By 1,2) immediately below I was using a different numbering from earlier. The reason for this is that I am now calling 1) the simulations based on 4BVH
receptor, which you are currently running. 2) refers to the "preparing as in case of 4FVT with loop replacement from 4BVG (with coproduct ligand instead of INT)" simulation
approach (called 1 earlier below).
RSK: Thanks for clarifying. I got confused with the numbers.
See the commentary on this alternative approach. I also mentioned the Fri before you left that we will decide which one to use after seeing your preliminary
results. The reason for this is quite simple: the energies obtained from use of 4FVT structure may be more comparable and consistent with those we currently report in the paper.
Use of 4BVH could introduce various significant differences rendering the results inconclusive. Hence we may not have a choice.
We need to review the single point calculations before deciding. Note that use of the OPLS force field may cause problems in comparing the results in this regard.
Not clear that it should require 5 business days. We can discuss how to speed it up further at that time if needed.
RSK: Okay, I got it. So you want me to model 4FVT ( sirt3/peptide with ADPr product) and do a side chain repacking of the residues around 7.5 Amg of the ADPr product. Similarly construct another model starting with 4FVT but the loop replaced/grafted from 4BVG and the product ADPr placed into the model followed by repacking of the side chains. In both the models the acetylated peptide portion (lysine) is converted to a decateylated peptide followed by a side chain repacking for residues around the Lysine residue of the peptide. Eventually, your idea is to compute MM/PBSA single snapshot energies of the refined complex and compare them. I hope that I got your idea correctly.
If that is the case then it can becompleted in 2-3 days.
RC: The structures can be prepared based on the starting structures for the MD simulations currently reported in the paper (4FVT w/, w/o loop replacement from 4BVG, with INT ligand placement, side chain prediction already done). INT would be converted to Lys-peptide and coproduct and side chains prediction around Lys would be done as you indicated above.
RSK: Okay, I got it. So
I believe that the 4FVT and 4BVG INT complex which we have in the current manuscript has NAM present in the complex. Do you suggest that we can use these (Sirt3/INT/NAM) starting structure for our new simulation (Sirt3/product without side chain modelling and prime refinement, but instead knock out NAM and convert the Acetylated Lys peptide to an Deacetylated peptide.
--"answers to the remaining questions in the latest question list..." point below should be addressed.
RSK: Answers to your latest questions.
computational tasks 6-7c_RSK .docx
Answers to the questions. I guess this is the one you asked for.
--side chain validation part should not be removed from the miscellaneous task list and schedule. If desired it could be put toward the end of the schedule.
RSK: Sure, will add it to the end of the schedule. In fact we have not collected much information on the validation data earlier..
Related, in the marked up side chain prediction doc, the side chain validation notes of Ping were mentioned.
The new protocol posted for this is a way of organizing the results from validation more concisely and systematically.
RSK: Okay I will start gathering all relevant data from Pings desktop to organize this section.
RSK (6/9): A revised schedule is updated.
revised_schedule_V3.docx
RC (6/13): Since we want to compare the Amber ff energies of 4BVH and 4FVT prepared coproduct structures (methods 1,2 mentioned above) before proceeding with production simulations,
we may consider doing equilibration for the prepared structures and then examining the energies after equilibration as an alternative to comparing single point energies. (I am assuming
most of the time required would be spent on setting up the Amber scoring so doing a single point calculation rather than equilibration would not save much time.)
Since there are two complexes that need to be prepared and scored for method 2), you can do a shorter equilibration for these complexes for this purpose, if desired.
RC (6/8):
-- Revised schedule didn't account for fact that there is one unfinished simulation and its scoring. Only the new simulation (open loop) is scheduled.
--Miscellaneous tasks list and order were not updated in schedule
RSK(6/9): Updated now. I am not sure of the side chain validation part and I think you will be giving me more instructions on this section later. Hence I have exclude it for the time being form the work schedule.
The latest version of the questions and misc tasks list doc (now updated once again, see below) were not used in the answers to questions. Priorities for miscellaneous tasks were also given there, along which were ready to start at any time
If 2 sims must run and schedule will be pushed back would like to see schedule for which misc tasks will be done during that time.
RSK(6/9): A revised schedule is uploaded based on the priory as listed by you and considering the time of availability of the data ( one simulation is running and the next one needs to be set up)
--Need to see mm gbsa energies from the closed loop sim imm after competition of sim . [If you have these results from PLs sim you can send those now, but I doubt you have them scored.]
RSK(6/9): Okay, that is now included in the revised . I have not computed the MM/PBSA values from Pings MD trajectory.
-- Following software installation, answer the remaining questions in the latest question list, including those pertaining to sidechain pred (sep document) before moving on to the structure preps.
-- As noted on wiki and during our discussion just prior to your trip, we would decide around this time which pair of simulations to run and report in the paper. Adjust schedule to do structure prep for both following methods before proceeding. You have scheduled 1, but I would like to do structure prep for 2 concurrently (before running new simulations), since I do not want identify inconsistencies only after the simulations. If 2) takes some time to prepare, you
can run simulation 1) in the meantime.
1) starting from coproduct complex xtal structure, one could replace loop with apo loop conformation (open). No peptide.
RSK(6/9): I think in the interest of time and the advantages Sirt3/AADPr complex with an closed and open loop conformation offers, we decided to go with this option over other option we had ( 1 and 2) (listed as 3 on the wiki ).
RSK: I am not clear by what you mean by “structure preparation for 2”. Sirt3/ADPr with closed loop (4BVH) does not require any modelling of the side chains or the loop region (Its just a simulation of the native structure after knocking out the inhibitor). Conversely, Sirt3/ADPr with an open loop requires modelling using Prime. I believe that we are considering only this pair for the current study. Please advise me if I have understood wrongly. This has been listed this in the schedule.
Report the single point MM-GBSA energies for starting structures (or energies from a v short MD simulation) from both open/closed for preliminary analysis. This should ideally be done with Amber ff for proper comparison.
RSK(6/9): Okay, but it can be done only after the simulation has been started for Sirt3/ADPr with open loop.
2) preparing as in case of 4FVT with loop replacement from 4BVG (with coproduct ligand instead of INT). See wiki for details how to prepare the structure, which includes removal of the acetyl group from Ac-Lys peptide to make it Lys peptide (includes minimization, analogously to INT ligand prep from the INT simulations). Possible advantage of this approach: energies may be more comparable to those already in the paper due to the use of the same global protein structures in both sets of simulations (INT and coproduct simulations), and peptide is present in both cases.
Note that loop conformations are somewhat arbitrary since we are concerned w modulation, so the slight differences between INT/coproduct and ternary/apo loops are not so significant.
For side chain opt, predict with the same method within the standard radius of the deacetylated lysine (for both the open and closed loop conformations), since the substituted 4BVG loop was already subjected to side chain opt in the corresponding INT simulation. You may then check the side chain RMSDs with respect to those in the starting structures for the INT open/closed loop simulations.
RSK(6/9): This task would consume 5 business days and if listed on the schedule it would push the deadline beyond June.Please let me know if this an priority. My understanding was that sirt3/ADPr with open and closed loop conformation were of utmost priority and we settled for it.
Report the single point MM-GBSA energies for both open/closed (we should have averages from MD simulation of closed by this time) for preliminary analysis.
RSK: A quick single point energy can be obtained from Prime Schrodinger.I think for single snapshot energies we can use Prime , because setting an calculation in Amber takes time.
After receiving the single point energies for 1,2 above we will decide which remaining simulations to run. As noted, if 2) takes some time to prepare, you may start simulation 1) after reporting the single point energies.
RC (6/5):
Vijayan,
Since I did not hear from you I am assuming you did not start the closed loop coproduct simulation yet. It appears that resolving the bottlenecks, launching the simulation and running the GB(PB)SA scoring will take at least 5 business days. Unless extra hours were put in this would then push your schedule into the fourth week of June, even not accounting for the additional days I stated would be required to finish the miscellaneous tasks (for which you allotted just 2-3 days). This assumes that the bottlenecks with software related to partial charge fitting will be resolved within a day or two at most, which may not be the case.
I have attached a document below that includes: a) a list of questions and comments regarding the tools you are using in the computational work vis-à-vis those used by Ping; b) a revised list of the miscellaneous tasks. Note that there were some miscellaneous tasks that were missing from your list and in addition, I have added a few new tasks. The miscellaneous tasks list also indicates which of the tasks can be done concurrently while the jobs run. I would like you to allocate these in parallel so you do not lose the full week. The remaining miscellaneous figures/tables will often be analogous to the ones that can be completed now, and I will be making edits that will affect all of them analogously. The miscellaneous tasks may shortly be split into two: a) mandatory; b) optional.
computational tasks 6-7c.docx
RSK: I will revise the schedule and answer your questions by today. In the interest of time and to clear the backlog, I am planing to edit the wiki after office hours today. is that okay with you?
RSK: Please find attached the revised schedule and my response to the computational task questions. I have used after office hours to edit the wiki and for compiling my responses considering the tight deadline for the paper.
computational tasks 6-5_rsk.docx
RSK: The revised schedule
revised_schedule_V2.docx
RC (6/6): The computational tasks doc above has been slightly edited to indicate which tasks are of lower priority (marked "optional"). They will still be done if time permits,
but the others will take priority. If it is natural to do some of the lower priority tasks concurrently with higher priority ones due to relationships/overlap between the tasks, it is fine to do so.
Next, I will be posting the marked up side chain prediction protocol. Following that and further progress, I will be posting a Methods outline.
It is ok to update after hours, as long as there are no bottlenecks that I should know about in fitting the charges and starting the simulation. I am assuming here that you just didn't get around to doing the work as per your email.
Note that the computational tasks document above has now also been edited to include a detailed side chain prediction validation protocol and format for presentation. As noted, some of those steps may be optional based on available time.
This protocol should be reviewed in conjunction with identification/summary of PL's previous work on side chain prediction validation for these enzymes.
RSK: I am not clear about the side chain prediction validation protocol section .I guess that this work has bee carried out Ping. Do you want me to look in to those data he has compiled.
RSK: My understanding is that we have not compiled any side chain validation data yet. Also, I have not come across documents from Ping describing the protocol. I have stumbled on some xls sheet but I never looked at them closely.
RC (6/5): Separately, I will also be sending you a marked up copy of your side chain prediction and refinement protocol document with comments on missing items/items that need revision vis-à-vis the work Ping and I previously did on this topic.
Side chain prediction and refinement protocol RC comments.docx
I understand that you may not be able to answer some questions regarding the previously used protocol for structure preparation without updating the license. You mentioned you need the license to conveniently review the log files.
If you are encountering issues with updating the license without reinstalling the software, please summarize them here.
RSK(6/9): My comments to the your questions contained in the above document "side chain prediction .....RC comments.docx"
Side chain prediction and refinement protocol RC comments _answers_rsk.docx
Also, I am preparing an outline of the Methods section [with subsection headers and comments on what needs to be included in each subsection]. I will send this after I receive your replies to the questions in the attachment(s) and we communicate on them. Note the questions include a request for an updated schedule as well as a summary of current status of bottlenecks with charge fitting. Please prioritize your answer to the latter asap.
RSK(5/27): A revised work schedule detailing the list of miscellaneous task that needs to be undertaken for the manuscript is provided . Unfortunately, most of the miscellaneous task cant be undertaken prior to the completion of MD work as they are tied with the MD results. Hence, it would be hard to push them to earlier dates. However, some items on the miscellaneous task list can be done as and when time permits even before the completion of MD work.
revised_schedule_V1.docx
RSK(5/27): A working draft for side chain prediction and refinement adapted based on Pings previous work. This is primary for our upcoming modelling of Sirt3/AADPr with an open loop conformation.
Side chain prediction and refinement protocol.docx
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
RSK(5/27): Attached is the revised schedule considering the tight deadline for the paper.
revised_schedule.docx
RC (5/25): Thanks for the preliminary replies. I will reply to your questions but after I receive your replies to remaining questions below, with emphasis on the following priorities:
- schedule refers to not only how long it takes to do each step of a protocol, but estimated dates on which you anticipate completing those steps of the work (this is esp important since you apparently may not be able to start some simulations and associated structure preparation until after you return from India). You may assume that simulation 3) without side chain optimization can be launched during your trip, but see below regarding caveats on this. It would be best to assume you will need to run another simulation including the full structure prep protocol after returning from your trip.
RSK (5/25): I have attached a detailed schedule that runs through entire June for completing all the structure preparation and other MD task for all systems you would like to consider.
Also, please correct me if you feel that my work schedule listing/or my explanations of computational aspects are too verbose. If you feel so then I can stick on to a high level description in the interest of your time.
Schedule_June.docx
RC: Thanks, I appreciate putting the schedule in format consistent with the formats we have used in the group in the past.
I will review and revert.
RC (5/25): I have briefly reviewed the above schedule and have several comments:
- I do not see a detailed schedule regarding the miscellaneous tasks that must be completed for the paper (some but not all of which were listed below)
other than an entry towards the end of June. As noted I was planning for those to start asap and concurrently with other work so I can finish various parts of the paper.
- The other simulations you mention would start in 2nd half of June are not all required for the paper. As noted we would choose among the three options below shortly.
- See new comments below regarding coproduct simulation without peptide. Peptide is not required for this simulation if it is not already there.
RSK: Okay, got it. I assume that we are just going with just two simulations here
1) Sirt3/2-OAADpr complex with its native loop ( which I am currently working )
2) Sirt3/2-OAADPr with loop grafted/replaced from apo enzyme (3GLS- A open loop conformation). This task involves prediction/repacking of side chains for a defined region followed by Prime energy refinement subsequent to the grafting/replacement of the loop.
Thus the two sets of simulations (with, without peptide) are not required. There is just one pair of simulations corresponding to #3 below., one of which you are currently working on.
RSK: Okay, I got it. We are not considering deacetlyaed peptide as a separate system. Just going by an single pair
Please revise.
-If we choose to go with method #3, there would be just one structure preparation followed by MD prep/simulation, MM-GB(PB)SA
analysis after you come back.
RSK: Yes, you are right. One MM/PBSA analysis followed by one structure preparation and MD simulation for that system.
- When you say you would launch the open/closed loop simulations (for #3) prior to your trip, which do you mean? You were already running one this week,
for which you were planning to run MM-GB(PB)SA next. Are you referring to restarting that given the modification to the partial charge assignment protocol?
Or are you referring to the open loop simulation? Both concurrently? Please indicate in schedule.
RSK: Yes I am referring to restarting the Sirt3/2-OAADPr native complex with Ex527 knocked out. I did my trial with AM1-BCC charges, however, for consistency with Ping's work I had to use QM charges. Unfortunately, I stumbled upon on many issues when I started executing the QM program. I had been trouble shooting and fixing each issues one by one mostly software related ( Mostly issues related to installion and libraries).
This is just an one time fix,we will not have to encounter this problems in future. I am still baffled how these issues arise. I believed executing the programs should rather be straightforward because, Ping had used it earlier.
RSK: I thought that I can set up one simulation with a open loop ( grafted form apo enzyme), but without side chain chain prediction. I now realize that this is not necessary, because we need one with structure prepared. Hence I removed it form my task list.
Ideally it will not take the rest of your time before your trip to launch the simulation(s), and if more time is needed perhaps you could launch the open loop simulation for #3
while on your trip since the other work will not likely be possible while traveling. If it is taking more time than anticipated please elaborate.
RSK: Right, since I am not doing any simulation or other tasks during vacation, I think I can set up the script for computing the MM/GBSA during the vacation so that I can get you values the day I return.
RSK: I remember that I assured you that I will be setting up this simulation early this week. But unexpected issues with the PC-GAMESS software (mentioned earlier) and the urgent need for coming up with work schedule and updating the wiki ( yesterday)holded me form launching the job yesterday..
-Note that the open loop simulation for #3 without side chain optimization may not be usable for the paper. Thus I was endeavoring to see some progress regarding the
various other tasks in the near future, and/or the structure preparation protocol, which can be used for the paper.
RSK: I got it. It will be removed form my task.
See questions posted regarding the structure prep protocol and docs, which appear to be pushed back to around June 10 (about two weeks from now).
I had been hoping for some organization of the shortlisted docs this week, so you would be ready to apply the protocol immediately after returning and
so I may do some further work on this while you are gone. Please look into this if possible.
RSK: Okay, I will look into it tomorrow, and come up with a clean draft for your approval. However, I have a question here. Can we be so rigid with a single protocol which Ping has used for modelling Sirt3/INT/NAM complex, without exploring the other options available on Prime. At times a method which works fine for a particular system may not work fine for the other ( I mean it in terms of energy, H bond network, Chi angle propensity ). I believe that Ping has explored all options and chosen the one that produced best results in terms of Prime energy ( least negative ). So could you please clarify If I need to explore all options and then settle with the one that produce the best ( in terms of energies) or straightaway use the option which Ping has identified to be better. The latter would be straight forward and could reduce the structure preparation task even by a day.
-The overall schedule of getting writeups of the results at the end of June and spending most of June almost exclusively on setting up MD simulations
is unfortunately not so consistent with the anticipated schedule for submission of the paper. The first two weeks of June would ideally be spent on filling in gaps
in the paper/finalizing paper sections while 1(-2) simulations and scoring calculations run. We want the paper to be ready for submission by the end of June.
Try to schedule in the miscellaneous tasks bearing this in mind.
RSK: Sure, I will come out with a new schedule. Considering that we are going with only one more structure preparation an MD, the schedule is definitely going to be well ahead and on expected lines with your time frame.
- You mentioned Schrodinger license server installation and uncertainties therein. This was already done in the past and the configuration worked out. I believe
there are notes on the wiki. Have you reviewed these? This should not take much time -- I believe Arabinda is familiar with it, and it should primarily be a matter of
using the newly provided license file.
RSK: I do remember coming across some documents on Schrodinger installation. If it had been an license upgrade it would have been very easy ( just replace the old license file with the new one and restart the license server). However, Schrodinger software has many new additions ( like OPLS 3), So to access these new functionalities we need to install the latest version and not just update the old license file. I completely agree that 2 days would be off the limits. This will be only in case of the worst case scenario.
I anticipate that installing the software on the windows will be relatively straight forward but ensuring that our GPU nodes catches the license from the Windows machine is the tricky one. In fact, I had done a similar installation between two Linux machines and I hope to replicate it between a windows to Linux machine this time.
--Overall the most important point is that I anticipated less time to completion of the structure preparation and simulation of the 1 (at most 2) complexes remaining to be studies for this paper,
and more concurrent work/more detailed schedule toward completing all the other pending tasks that are necessary for paper completion (including some progress this week on the latter).
RSK: Sure, I will take it in to consideration this aspect during revision.
A schedule that ends in June would most likely push the paper submission back to the second half of July.
I will review more carfeully, elaborate on some of the above shortly and may revise the schedule after you edit it in response to some comments above.
We can discuss thereafter if needed since I acknowledge the ideal schedule may not be very feasible and I may be oversimplifying some of the above.
I am considering allocating at least some of the new tasks regarding image editing to other lab members; let me know whether this would be advisable given your schedule.
- with respect to structure preparation protocol, underlined comments were priorities: including providing a short list of only those of Ping's documents that are
required for/used for the results reported in this paper.
RSK (5/25): Regarding image editing my opinion would be that you can list it as a common task. I can always step in and volunteer whenever I find time. I can co-ordinate and work closely with other members on these images.
The final choice of simulation(s) to run will be determined (and further clarification provided) after receiving all the requested background info.
RC (5/23): Continuing from post below on "Considerations in the choice of structures for use in the coproduct simulations:"
Noting that both open/closed loop conformations are required for coproduct, three options presented below were:
1) preparing as in case of 4FVT with loop replacement from 4BVG (with coproduct ligand instead of INT)
-# new simulations required: 2 (a,b)
-structure preparation required: minimal, since loop replacement and side chain opt already done (under approximation that further side chain optimization due to change of ligand is not critical).
replace NAD+ in 4FVT with AADPR and ac-peptide with deac-peptide, rather than replacing with INT
-rigor: lowest
RSK (5/24): My understanding is that I should prepare a system starting from 4FVT but the NAD+ in 4FVT should be replaced with AADP and the ac-peptide should be replaced with deac-peptide. Similarly, another system starting from 4FVT with the loop grafted form 4BVG and the carba-NAD in 4FVT should be replaced with AADP and the ac-peptide should be replaced with deac-peptide. Let me know if I understood it correctly.
It can be done but would require side chain prediction with prime for at least one residue in the peptide and its close neighbors in the protein (sirt3) using Prime, because the ac-peptide will clash with AADP.
RC: Yes this is roughly correct. Given that 4BVH does not include peptide, it appears that one advantage of this approach compared to 3) is that peptide need not be placed.
You could place coproduct coordinates via alignment of 4BVH.
Another possible advantage of this approach, that will be considered further, is that the energies may be more comparable to those already in the paper due to the use of the same global protein structures in both sets of simulations (INT and coproduct simulations).
Depending on how long it takes to set up and carry out the structure preparation steps for 3) below, we may thus consider whether one of these simulations 1a,b) could be run concurrently with structure preparation (following local side chain optimization).
2) could alternatively start from apoenzyme xtal structure and replace with a closed coproduct binding loop, adding ligands
-# new simulations required: 2 (a,b)
-structure preparation required: high, since loop replacement and structure side chain prep must be redone for 4BVH loop
analogous to 4FVT/4BVG loop replacement, but with 3GLS/4BVH or similar for open/closed loops
lack of peptide substrate in 3GLS may cause problems and render this less option less attractive than 3) (this could be used as a justification for the use of 3)
-rigor: high
RSK(5/24): I believe when you say replacement with co-products here you mean both deac-peptide and AADPr.
If I use the 3GLS as my template and graft/ thread the 4BVH (product complex with closed loop) loop onto it, would cause problem as you had indicated. The reason being the structure of apo (3GLS) differs from product complex (4BVH) not just in the co-factor loop region, but globally. Secondly, placing/docking the substrate peptide onto 3GLS (apo- open loop conformation) structure from 4FVT or from any other peptide substrate complex as our reference would be tricky becuse it would clash with the peptide for reasons you had pointed out. I agree that it would be a less attractive option.
RC: I was not aware that the 4BVH structure does not have deac-peptide ligand. Still it seems the global structure of 3GLS (apo) differs significantly due to lack of AADPR, so the conclusion is effectively similar - 3) may be a better option.
3) alternatively, starting from coproduct complex xtal structure, one could replace loop with apo loop conformation (open). But this would be more analogous to the use of 4BVG with
loop replacement from 4FVT, which we are not reporting in this paper (see note under 2 above regarding why 3 may nonetheless be preferred).
-# new simulations required: 1 (1 already being done this week)
-structure preparation required: moderate, since loop replacement and structure side chain prep must be redone for 3GLS loop
-rigor: high
RSK(5/24): You indicate here that the other alternative could be using 4BVH (product complex closed loop) as the template and replace the loop region with an open loop conformation of the apo enzyme (3GLS) , together with the placement of deac-peptide ( ie- sirrt3-open loop/AADPr/deac-peptide) . I agree that it would be to carrying out a simulation using 4BVG (intermediate complex) as the global structure is nearly the same. However, in 4BVH (product complex) we see a small helical turn, on the contrary we see a completely unwinded loop in 4BVG (Intermediate complex).
I agree with your viewpoint that this may be preferred over option 2 ( I don't foresee an need for side chain prediction here so we need not wait for Prime license. I may be able to set up this simulation before Friday ( will try my best to set it up if not I also attempt my best to set it up during my vacation period) so that I will have the trajectory when I come back.
RC: I didn't mean to place deac-peptide if it isn't already there. So, you have already been running the closed loop simulation in this case.
Regarding side chain prediction, in the past we have not only applied it when we substitute a new ligand, but also for loop replacements. See for example the case of 4BVG with loop replacement from 4FVT, which is quite analogous to this simulation. Thus, in this case we would carry out side chain optimization upon substitution of the open loop conformation from 3GLS. So, I am not sure whether we can get away with omitting side chain optimization, if we are to be consistent. Of course, Ping's side chain prediction validation and application results in the non-native environments are relevant in this context. I cannot comment a priori about the extent to which side chain optimization was critical for the other loop replacements we did.
For the purpose of preliminary planning we will assume this will be set up during your vacation period.
RSK(5/24): Indeed the method/protocol what you have pointed out is right and I would say to be the best praticse. I completely agree that side chains have to predicted/repacked even after grafting the loop from another structure. I have heard some in the modelling community with a different viewpoint that why bother about side chain prediction if you are eventually planing to do an MD simulation which will refine the complex itself as it samples the side chain conformations (rotamers). Their argument that MD will refine the side chain conformations during the course of simulation would be only valid if we simulate longer time scales. However, for consistency with Pings works and considering the fact that our simulation are going to be short , I agree that side chain is necessary even after loop replacement.
RSK: Please let me if I understood the different systems and the method of preparation you had listed above ( 1 to 3) correctly ( I need to ensure that we are on the same page before I start preparing the system , which you will prioritize)
Further pros/cons and details of the approaches were already mentioned below.
-- We will finalize the decision on which (1-3) to use based on your estimated schedule requested below, in particular when we anticipate finishing all the required simulations for the paper
(under the assumption that only 1 new MD simulation will be done). If this is not too late, we will consider the other options 1,2 as well.
RSK: Time frame for each study
Side chain prediction and modelling: Side chain prediction and preparation (opls ff minimization followed by prime minimization using implicit solvent model for each system ideally can be done in a day or maximum 2 days (if some unexpected issues arise that needs trouble shooting). Applicable only if required (for some systems not all systems)
MD set up. We need to parametrize the non-standard residues if any like AADP and obtain QM based partial charges for them. This would require half a day to 1 day of work for each system. Also we need to derive special force field parameters for the Zn++ ion coordinated to four histidines. I am treating these coordinate bonds in a special way consistent with the protocol recommended in Amber tutorials.
MD ( first stage ): Before launching production run we need to minimize and equilibrate ( 200 ps of equilibration) the system and we need to check for the stability of the system ( time vs RMSD, time vs potential energy, time vs pressure, time vs temp) to ensure that the system is stable before launching the production run (15 ns). This work would require 1-2 days inclusive of time required for a slow heating up (from 15 k to 300 K with a gradual increase of 15 K) of the system and then running equilibration simulation for 200 ps at 300K.
MD (final- production run) – given the current scalability (~ 5.5 ns per day), ideally the job should be done in 3 days’ time, assuming that no other MD job are running on the GPU node.( The maximum run time will be a week (5 days) assuming we have 2 MD jobs running concurrently ).
MD (analysis and free energy calculation) – A quick check for convergence (RMSD vs time) and ensure the system was stable during the course of simulation. Then proceed to extract the trajectories and strip the water molecules from the MD trajectories and prepare it for an MM/PBSA and MM/GBSA re-run.
Setting up for an MM/PBSA re-run and re-running the trajectories for extracting binding energies/ conformational energies will take 1-2 days (I see that the PB calculations alone take about 8 hrs of compute time.
Ideally a single calculation (preparation/MD/MMPBSA) would take a complete 2 week time. However, when one process is running, I can use that time for setting up preparing other structures and the other simulation.
RSK(5/23): Slightly tangential, but for you information. This is related to the simulation ( sirt3/2-OADPr) with its native loop. The simulation will be launched today, I attempted launching one yesterday night form my home . I see some issues cropping up in generating RESP charges, anyways it sounds to be a trivial issue ( I need to set up a scratch directly it seems, that is the error message I get) it will be fixed by today. The trial simulation which I successfully ran and reported earlier was using AM1-BCC charges. But for the final simulation I am using QM derived partial charges, because I see that the PLOS One paper does mention RESP charges.
RC (5-20):
n Please provide an update on the time required for the 15 ns MD simulation if you have it.
n What is the status of the simulation?
RC: I note you have just emailed be about this while I prepared this post, please post it here.RSK: I am done with the trial simulation (Since this is the first time I am using NAMD I first set up a trial simulation to ensure confidence). Also, the trial simulation was necessary so that I can get an idea of how much GPU acceleration could be achieved on the pmcat-gpu1 hardware.
RSK: Looking through the command history files provided to me by Arabinda, and the ones lying on the pmcat-gpu1 node , I see that Ping has used only a single GPU device and 4 threads for his simulation.
However, I find that we can obtain better scalability, if we use the maximum available threads and multiple GPUs
The maximum available threads on pmcat-gpu1 are 22. It was determined as follows
Total number of physical CPUs = 24
Total no of GPUs = 2
Maximum threads = (Total CPU Cores – Total GPUs) = 22 threads
RSK: Using 22 threads for my simulation and I achieve about ~ 6ns a days for Sirt3 -2OADPr product complex. I definitely see enhanced acceleration (based on simple interpretation using short runs) using multiple GPUs. However I didn’t want to spend time in doing a benchmark exercise, given it less priority.
RC (5/23): We will most likely buy another gpu node for the purpose of our next paper. After you come back from your trip please request a quote for a node from Exxact for approximately the same price as the last one, with specs adjusted appropriately to match the price (not before, they will bother you and it will take up too much time at this point). If you don’t have the old quotes and specs, Wei can provide them to you. dd
RSK(5/23): Sure, I will touch base with Wei on this after I return back from my India trip. Also, I do have a suggestion here which you could give a thought. Why don't we go for MD packages like Ace MD ( the fasted MD engine for GPUs ) which are available via Amazon cloud. I believe that slowly migrating to cloud computing would reduce recurring overhead charges, time and money involved for system maintenance/ need for an system administrator, etc etc.. instead of investing money on GPUs.
Copied below is an link of one such company namely Acellera. You may have a look into it when you find time.
https://www.acellera.com/products/acecloud-molecular-dynamics-cloud-computing/
I think that there will be more vendors too. if you are interested, then I can looking more closely and gather information.
RC: I have already considered gpu computing on the cloud and Ping did a review of the options. I had suggested it at the outset and delayed purchase of gpu.
In fact, I already use amazon cloud for other technology projects at AT (you may note this from some of Arabinda's tasks; we host another wiki entirely for that work),
and have colleagues at companies that do MD simulations on cloud routinely. As I recall, Ping's conclusion was that it was not cost effective for us. You would need to review his notes at the appropriate time (not right now).
I still have a plan to implement it, but not without buying one more gpu node in the short term.
I did not mention it now since I did not want to slow down the immediate simulations.
As noted the quote task is only to be done after you return.
RSK: I will be carrying out some quality checks ( testing the intergity of the simulation) the trajectory to ensure if everything went with the simulation (RMSD vs time, Pressure vs time, Potential Energy vs Time).
RSK: I will be setting up a 15 ns simulation on Monday and I expect to get you the MM/PBSA and MM/GBSA data by end of next week
n I had asked for a list of the structure preparation documents from Ping that you have collected which you deem relevant to the preparation of the starting structures for the simulations that have been reported in the paper.
Given that the simulation you are currently running (without structure preparation) is not sufficient for the other loop conformation (open), which may require analogous structure preparation, we need some lead time to review those documents and plan for the next simulation (according to the notes posted below on the different approaches to setting up the remaining simulation(s).
RSK: A short summary based on my understanding after going over all the reports is provided here.
1) The initial protein structures were prepared using the “protein preparation wizard” of Schrodinger. This is for assigning protonation state, bond order and for adding hydrogen's.
2) He has used OPLS ff for initial minimization.
3) For final refinement ( after loop refinement and side chain prediction) he has used all-atom OPLS force field and VSGB 2.0 solvation model.
4) For side chain prediction he has used the Prime Monte Carlo method (MC) method with backbone sampling or CA-CB vector sampling in some cases.
5) He does mention in one of the documents that “Using Monte Carlo method in 3-residue side chain prediction usually lead to slightly lower energies"
6) He also states that he saved the top 8 conformations provided by the MC run, but used only the “top few” I guess 3 for prime minimization (OPLS ff with VSGB foie electrostatics and solvation).
7) He has followed the default with 3 residues optimization approach ( That includes 3 extra residues for optimization beyond the defined residue or region of side chain prediction).
8) For loop modelling he has used the rigorous “ultra-extended sampling” protocol available in Prime
RC (5/23): Ok. Does this include methods for nam, int (ligand) placement for old sims?
RC: Please answer above.
Regarding the structure prep protocol, based on your review of these docs are you comfortable applying the same protocol Ping did?
RSK(5/23): I am comfortable with the side chain prediction and loop modelling protocols implemented by Ping. I may need a couple of days to familiarize with the "extended loop sampling protocol", which I am familiar with but have not implemented it in real time in any project. Further Prime loop modelling is mostly through GUI and much easier compared to its academic variant PLOP.
RC: As noted we are not applying loop prediction in this paper.
My question here is, are you ready to implement (and describe) the exact protocol that was used by Ping for the other MD simulations reported in this paper, not just a preliminary outline?
The writeup itself will be scheduled, but I need to know whether you are in a position to implement and write up whenever I propose the simulations to be done.
It was my understanding that many if not all the steps were clearly described in some of the reports. If so, why do we refer to other issues like loop modeling?
You should indicate a short list of only those docs from Ping that are relevant to only those results reported in the paper draft.
If you are not clear on the protocol, let me know asap and I will review the docs and help clarify. Note that I have worked with Ping on steps above and can advise if needed.
See also below regarding side chain prediction validation (somewhat lower priority).
Use of gui is convenient and I am aware of that, but in general it is not a reason to not be familiar with command line implementation. E.g., extended loop sampling (which we are not using here) requires knowledge of the input file commands. See the previous correspondence with Ping on this. You should be able to run from command line so whenever we want to use our own source codes we can without delay.
Once I receive confirmation that you are able to replicate Ping’s structure preparation protocol including loop replacement, I will advise on which simulations to set up, based on the considerations I outlined below.
We will consider the schedule for this work thereafter. Given the estimated time per simulation, it appears important to settle the plan for when the structure preparation protocol will be applied to set up the new coproduct simulation including structure prep that we will run next. I would like to determine whether or not we should wait for the schrodinger license to start before implementing any of the steps in this protocol, since if we do wait, it appears we will not be able to run the simulations while you are away. In that case, we will consider whether there are any other coproduct simulations that do not require structure prep that can be run while you are away. Alternatively, we could use plop or pre-pone the start of the schrodinger license.
RSK(5/23): I agree that we should schedule it for a later date and wait until we get Schrodinger installed and get it working. I believe that is not too far, we will be getting the license in the first week of June and that would be the perfect time, because the other miscellaneous tasks would have been complete by that time and have an idea of what method should be employed for modelling the open loop conformation.
RC: Are you saying the structure preparation as well as MD simulations should all wait until after you return from trip? I need to schedule this since
the simulations will take time and it looks like the paper may be pushed back a month due to these issues.
No simulations will be running while you are out?
Estimated schedule for the pending structure prep implementation and simulations should be provided. For this purpose assume there will be one more coproduct simulation
with open loop conformation that requires structure preparation.
RSK(5/23): I think that the other co-product simulations (open loop, intermediate loop ) will certainly require loop modelling and structure preparation steps.
RC: We are not planning to do any loop prediction in this paper, unless there are missing loop segments. Structure preparation including loop replacement and side chain prediction will be used.
Please note that I have another paper draft in preparation that is longer and involves other loop simulations. I have not shared this yet since getting this paper out is the priority.
RSK: The list of documents which I picked up form Pings desktop system, that would be of interest for side chain and loop modeling are contained in the txt file attached below
list.txt
RSK: All the relevant files listed in the text file is dumped into this zipped folder attached below.
Loop_documents_Ping.zip
I have also shared a copy of the folder via dropbox. In case wiki has issues with download you can access it from Dropbox.
RC (5/23): Regarding structure preparation, as noted there was some work done on side chain prediction validation by Ping. Recall we also discussed this topic during your visit. In the SI we may report on some of Ping’s validation results. We will decide whether to present them after you review the validation procedure and are comfortable organizing the results.
Since for this paper, we will not be redoing old sims, we cannot change struct prep method. We will continue to develop it for the next paper. Correction of energy or sampling errors will also not be considered in this paper.
RC: Please also provide an update on the status of the remaining tasks listed on the wiki, in particular which other pending tasks you are currently working on while the simulation is running.
Based on the status of your work I will determine when to assign you some image editing tasks, which may be soon.
RSK: I will provide you a complete list of pending tasks if any on Monday.
RC (5/23): Since the simulation you are currently running will take all wk, please report on status of other miscellaneous tasks (incl ligand interaction diagrams and pdfs) and the associated schedule for those. The schedule is esp important since I believe you will be out of town next week. After I receive this schedule I will as noted incorporate into that schedule some image editing tasks as well.
RSK: I reviewed the wiki page and I find that the pending items on the task list are
1) Obtain probability density distribution (pdf) plots for conformational energies and time series plots for MM-GB(PB)SA energies of MD trajectories
2) 2D ligand protein interaction diagram using Ligplot/or Schrodinger tool.
3) A more comprehensive write up of the side chain/loop prediction protocol based on all the documents gathered from Pings documentation.
4) Computing the standard error mean (SEM values for the MM/PBSA and MM/GBSA values which I re-ran ( every 1 ns time interval)
RC: The schedule for these is imperative. Schedule whichever ones you can for this week and specify the order. The above are not necessarily in order.
I am going to add another image editing task now since I have not yet received the schedule. If any additional preparatory steps are required before you can finish 1,2 I would like you to simultaneously
work on 4 so as to provide that promptly and paper draft preparation can continue (also, I realize that only a couple of the ligand interaction diagrams can be finished at this time).
I need to know when you will be able to finish 3 so I can further advise on the remaining simulations to run -- see above.
No loop prediction only loop replacement.
I also did not see replies regarding B factors and possible revised choice of reference for the Sir2 alignment mentioned below. Keep a list of all misc SI tasks so none are lost.
RSK (5-18):
I did set up an trial MD simulation of Sirt3/2-OADPr (product complex) by removing the inhibitor Ex-243 form the native crystal. The simulation ( equilibration step ) in NAMD is currently running fine ( The system didn’t explode or no other issues).
I will be running a short 4 ns production simulation today and then I plan to set up a 15 ns second simulation for our MM/PBSA and MM/GBSA calculation by tomorrow. Will provide you a time estimate of how much CPU time would a 15 ns simulation would consume by tomorrow.
Also, I have installed GIMP , the image editing tools which Guan and Ashok had been using for editing images.
RSK(5/13/2016) : A short status report on work progress
1) I am almost done learning setting up NAMD simulation using AMBER ff parameters. I hope to launch a trial simulation by Monday.
2) I have located all the loop/side chain modelling report prepared by Ping as requested by you. However, I need to read it more carefully and come up with a write up to make it more cogent, so that it could be eventually used for writing the method section of the paper.
RC: It would help if, as time permits, you make note as to which documents you are considering to be relevant to the protocol so I can verify and if needed, provide further input.
3) I have revised the supplementary section for the document which you had placed in dropbox.
RC: Ok, I will take a look. In the RMSD plot (and perhaps elsewhere in figs, have not checked), Sir2 may be referred to as Sirt2.
Also, in this figure, the RMSD is calculated with respect to the ternary complex, whereas for SIRT3 it is calculated with respect to the intermediate complex.
These references (which I believe were made by Ping originally) are not consistent. They Sir2 plot should be calculated with respect to INT complex. Also, the numbering is wrong for Sir2.
Note that the apo loop conformation in this Sir2 apparently also has a significant RMSD with respect to ternary (this may be consistent with it being referred to as open and ternary closed in literature).
If we show this plot we may need to also show a xtal B factor plot for Sir2 loops demonstrating that the apo conformation is more flexible. Also, the binary (peptide) complex should be replaced in this plot
with the coproduct complex if available. Please note that for the PLOS paper, Ping did many simulations with Sir2 in addition to SIRT3. So we may have data on file regarding those complexes as well.
RSK: Indeed you are right. Thee data for the plot was taken for an xls document which Ping had prepared and I re-plotted it. Looking at all the available structures in PDB , I see that we do have the complete loop structure resolved for Sirt2 apo (2H2I) and Sirt2 deacetylated p53 peptide-3'-o-acetyl ADP ribose (product complex- PDB ID 2H59). However, we don’t have the complete density for the co-factor loop region in Sir2/p53 peptide/NAD+ ternary (substrate) complex (PDB id: 2H4F) and Sir2/S-alkylamidate complex (PDB id: 3D81). Hence, if we useSir2/S-alkylamidate complex (PDB id: 3D81), which has missing density as our reference structure , we end up with a plot that has many missing data points because we cannot obtain data (residue-residue comparison) even for structures completely resolved. I am not sure if that is what you wanted.(please have a look at the figure embedded within the word document attached below)
I think that we may not be in a position to use an intermediate and substrate complex as reference here, because they have missing densities. This plot does incorporate all other charges you had asked for. Please let me know your thoughts on it. If this plot is okay, I will then go ahead and revise the SI draft which you shared via Dropbox.
Revised_sirt2loops.docx
RC (5/23): Ok. I haven’t carefully reviewed yet, but what about using Apo as the reference structure for both? Also, this is Sir2 not Sirt2 right?
Related, regarding the xtal structure alignments in the SI, in addition to the 4FVT/4BVG alignment we may also include such alignments for any other pair on which we report MD/MM-GB(PB)SA results (e.g., open/closed loop conformations for coproduct complex). This is of lower priority than the other tasks listed.
The comment about zooming in on the loop in the sequence alignment was due to fact that figure may not be readable at standard figure size. However, in SI the figures are bigger so it is ok for now.
I have in the meantime been making a few changes to the SI and I will assign some new tasks based on our most recent versions.
Also, please let me know once you have installed the image editing tools used by Alok and Guan to generate the various figures in the draft, and identified
the image files in the dropbox folder. I will then be assigning a couple of additional figure editing tasks.
4) There are two figures that have been left out form the supplementary data, which I had pointed out in the wiki document. All comments are embedded in the document. They can be tracked enabling the “track changes option”
RC: Yes, regarding the 2d ligand interaction diagrams, I have drafted a couple of additional figure captions that we will fill in with the plots depending on which coproduct simulations we do. I also indicated
that we may consider whether to show the starting structures for MD simulations (following structure preparation); this is not critical, and we may decide after looking at one example during finalization of the structure preparation protocol for methods.
We had both also referred to pdf plots at end of document to replace the convergence table. Regarding standard error of mean, no changes are needed at this time; we will remove this table when ready with the plots.
I have placed R2 in dropbox, removing the comments that we have now resolved.
You will note that I have also reordered some of the figs.
RSK: I see this as the only pending item on the task list. ( I will work on it over the next week as I will have more time next week when the simulation is running).
This work got side tracked as the priority task was on setting up the MD simulation this week.
5) I have already a product complex (Sirt3/2O-ADPR) by knocking out the Ex-542 form the original crystal structure. I hope to launch the production MD simulation job by Monday if everything goes fine.
RC: How long do you anticipate it will take to run?
6) A proposed second modelling approach using the intermediate loop conformation, will be discussed in detail with you before I undertake the modelling exercise.
(I hope that MPI and NAMD will work fine on the gpu node. I will be testing it on Monday. Also, I am not sure if Ping has done some benchmark calculations regarding the scalability of NAMD in the GPU cluster. In general NAMD program are scalable to a large extent. I believe that I can take advantage of all the 24 physical CPUs on the pmcat-gpu node. )
RC: Ok, I believe we discussed that you would be looking into this as time permits.
RC (5/9)
--Questions/comments on the INT binding affinity calculations:
-what receptor did you use (with or without NAM)? we had discussed running calculations with both. is one pending? provide data for the other one when ready.
RSK: I defined NAM as a part of receptor ( Sirt3+ NAM ) the ligand to be (INT) and the Complex to be ( Sirt3/INT/NAM)
RSK: Regarding defining sirt3 alone as as a receptor (excluding NAM from receptor) , I am bit skeptical because the numbers we get from it may not be reliable. The explanation is provide below.
We are following a "single trajectory" MM/PBSA approach in our study. ie, we are not running three independent simulations ( receptor separately, complex separately and the ligand in water separately), on the contrary we are splitting a "single trajectory" in to three separate trajectories assuming there will be no major conformational rearrangements in the receptor upon ligand binding and the strain energy of the ligand upon transformation from an unbound to the bound state is negligible here.
In fact this approach ideally couldn't account for the "reorganization energy" of the receptor and the ligand in the delta G calculation. This approach is usually advocated for estimating the "relative binding affinity" of congeneric ligands assuming that each ligand contributes equally to the "reorganization energy " and hence the they cancels out.
The problem here is our complex will be ( Sirt3/INT/NAM) and our ligand will be (INT) and the receptor will be (Sirt3). In our case then the complex will have 3 species, the receptor 1 species and the ligand 1 species ( the number of species in the receptor and the ligand don't add up to the total number of species in the complex). Hence , what happens when we plug the values form an MMPBSA run into the equation Delta G = Ecomplex - Ereceptor - Eligand,
a) the internal energy terms (bond, angle. and dihedral) which has to CANCEL OUT because the conformation of the receptor and the ligand are unchanged in bound and unbound state, will not happen in such scenario.
In other words the internal energies of the receptor and the ligand will sum up to the Internal energy of the complex, because the coordinates of the receptor and the liagnd are the same in both bound and unbound state.
b) not only the internal energies, the other non-bonded terms will also be different because the Ecomplex will have other non-bonde contributions from NAM as a part of the part of the complex but not in the receptor. This certainly brings some inconsistency.
RSK: Please advise me if my apprehensions is valid and should I go ahead and try running it.
RC: I understand there are significant concerns and I have considered them as well, but it was not clear from original posting what was reported in the document, since we had discussed both and you had inquired which should be run.
We had agreed the one you ran is the priority for the reasons discussed.
Since it appeared to take no more human time to run the other, I thought you were planning to run it as well and then analyze the data thereafter bearing in mind the reservations.
If it takes any time at all on your part to run it, you can postpone. As a matter of fact, it is not worth discussing it further at this time.
I have not prioritized analysis of how this data would be used at this time given the numerous other tasks at hand.
My main point was to establish whether you ran the right calculation.
RSK: Okay, I got it. I did run the correct calculations. These calculation took about 4.5 hrs to complete. Then I wrote a simple shell script to automate the process of extracting the energies. Okay, I will postpone it for the moment.
I am moving over to address other tasks that include protocol for structure preparation , Supplementary file on the dropbox folder , and the probability density function plot.
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
-please report standard errors of estimates
RSK: Okay, I do have the values,I will incorporate it into the report.
-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
-please confirm you used the same protein coords from 4FVT for both (i.e., only loop from 4BVG), when indicating modeling was done based on 4FVT or 4BVG.
RSK: I used the same trajectories used by Ping. Both the simulation had the receptor coordinates were taken form 4FVT but they differed only in the loop conformation.
RC: Ok good, I had asked because Ping also ran simulations based on 4BVG with loop replacement (you probably saw that data).
ie) Simulation 1 with Sirt3/INT/NAM modeled from 4FVT (ternary complex)
Simulation 2 with Sirt3/INT/NAM model from 4FVT (ternary complex) but the co-factor loops replaced using 4BVG (native intermediate)
RC: --For coproduct calculations:
-first identify all structure prep methods used by Ping for these particular simulations as systematically as possible. see all the notes/reports/task lists from Ping's work and
correspondence with us on this topic. It will be necessary to organize these for the purpose of methods section anyway as previously noted.
Note, e.g., that there were reports on the side chain optimization used after loop replacement.
RSK: I have identified all the relevant documents regarding loop and side chain modelling from Pings desktop and have them dumped then into a folder. If needed I can share that folder for your reference. I am going through the documents. Once, I get an clear idea after reading all the documents, I will summarize the method.
RC: -note the structure preparation will be somewhat easier in this case because there will be no NAM.
RSK: I agree that the structure preparation step is straight forward here. I believe that can easily model Sirt3/2O-ADPr product complex by knocking out Ex-243 for the Sirt3/2O-ADPr/Ex-243 (PDB if 4BVH). Today I also happened to compare the structure of human Sirt2/ADPr and Sirt2/ADPr/Ex-243 crystal structures. It shows that the binding of Ex-243 to the C pocket does not cause any structural change to the structure or the co-factor loop region.
RC: Ok, if we present simulation results based on 4BVH, we may consider showing such an alignment in the SI to demonstrate that Ex-243 doesn't induce such structural changes.
-Otherwise the structure preparation steps would be quite analogous to that used for the INT:NAM simulations, given the similarity between the INT and coproduct loop conformations.
-Note that in the case of INT:NAM simulations, we also did simulations based on 4BVG protein coordinates (with loop replacement from 4FVT), and this is somewhat more natural because the intermediate is the ligand
in the 4BVG xtal structure. We could do the same here if we like (or do both).
RSK: We can. Although structural evidences point that a "fully closed co-factor loop conformation" will stabilize the product, we can certainly try another simulation with an intermediate/closed conformation. My definition of the loop conformation for understanding purpose is
open conformation [seen in apo enzyme and substrate complex 4FVT( Sirt3/Ac-Cs2/Carba-NAD)] ( note the position of Phe 157 is different )
closed conformation (seen in alkylimidate intermediate - 4BVG)
fully closed conformation (seen in human Sirt3/2O-ADPr/Ex-527 complex) a mimic of product complex.
-i assume the coproduct coords from the complex with Ex-527 can be used for coproduct w INT loop.
RSK: yes it can be used. I don't see any major difference in the loop conformation and in particular the Phe 157 position between Sirt3 intermediate complex and Sirt3 product complex in the presence of Ex-243, expect for some change in helical content.
Attached a figure to illustrate it. Note the difference in the orientation the ribose moiety part of the NAD+
-before proceeding with any structure preparation, the protocol should be settled.
RSK: I will be launching a trial simulation on Monday in NAMD for Sirt3 product complex using the crusyal structure of Sirt3/2O-ADPr/Ex-527. The modeling here is pretty straight forward , I removed the Ex-527 from the complex.
RSK. Later, next week. I will start another simulation using the "INTERMEDIATE" complex loop conformation. I do plan to model it by grafting the coordinates for the co-factor loop residues from 4BVG onto 4BVH after appropriate structural superposition. I think that side chain prediction may not be warranted here, because they will no steric clashes ( not sure??) and then MD will do the job of sampling the side chain conformations. Just an idea. Please advise me on this if this protocol makes sense to you.
RC: I agree that there may not be much need for applying our full structure preparation protocol if we model the INT loop conformation with coproduct.
However, I am not sure the simulation you proposed above will suit our needs. See my bullet points below posted on Fri.
Briefly, if we do a simulation with INT loop conformation and coproduct, the purpose would be to make use of the previous structure preparation done for the ternary and INT loop conformation simulations already reported in the paper.
In that case, one can graft the coproduct coordinates from 4BVH onto 4FVT after loop replacement from 4BVG. Since the structure prep protocol was already applied to 4FVT with loop and INT coordinates taken from 4BVG,
it may not be necessary to do additional structure preparation if the INT coordinates are replaced by coproduct coordinates. If we proceeded this way, we would also use the ternary open loop conformation from 4FVT together with
coproduct for the second simulation.
On the other hand, if we used the apo loop conformation from 3GLS for the open conformation and 4BVH for the closed loop conformation, it would be a more rigorous way to model the coproduct complexes, but on the other hand would require more structure preparation
for the open loop (see my notes from Fri below for further discussion of the various issues in case this brief summary is not clear).
RSK: I looked at the structures of Sirt3/peptide/NAD, Sirt3/Intermediate, Sirt3/ 2OacADPr/Ex-243 and Sirt2–ADPR.
The intermediate complex has lost the helical structure, but apo sirt3, sirt3 /peptide/NADP and Sirt3/product/Ex-243 (or) Sirt2/product analog (ADPR) all have the short helix intact.
RC: Yes I recall seeing a short helix in some of your earlier posted images.
I was suggesting use of Sirt3/ 2OacADPr/Ex-243 when I referred to coproduct coordinates above.
Are you referring to Sirt2 or Sir2 - for which one is coproduct structure available? The former is mammalian, the latter is not. We will need to verify that references to Sir2 in the paper specify the right one -- the references therein so far are to the non-mammalian sirtuin.
You can prepare the Sir(t)2 structure alignment with coproduct for the SI (see prior note on this) based on your analysis when you have time.
RSK: I was refereeing to human Sirt2. I also remember reading that Sir2 also has a co-product complex structure resolved. I will check if the references to the Sir2 in the paper is correct and will prepare a structural alignment figure of sirt2 with co-product later.
Anyways, I am trying to set up simulation in NAMD for learning purpose, before we formalize the starting model for Sirt3/product complex..
RSK: Based on this observations, I think that we can remove Ex-243 from the Sirt3/ 2OacADPr/Ex-243 and use it as the starting conformation for our Sirt3/2OacADPr product complex simulation. Comparing the crystal structure of Sirt2/ADPR, which is a product analogue complexed to Sirt2 also reveal a similar loop conformation as seen in Sirt3/ 2OacADPr/Ex-243 (4BVH).
However, if one compares Sirt3/peptide/NAD (ternary complex) with Sirt3/ 2OacADPr/Ex-243 (4BVH) or Sirt2/ADPR (3ZGV), we see a short helix in the loop region in all three structures, but the difference here being the co-factor loop is more closed more onto the active site when it transition from an substrate (ternary complex) state to an product state.
RSK. The other alternative would be to consider Sirt3/ 2OacADPr/Ex-243 (4BVH) with its native loop and with loop region alone replaced from Sirt2/ADPR. In Sirt3/2OacADPr/Ex-243 (4BVH) we see a short alpha turn but in Sirt2/ADPR, we see a long helix.
RC: I will comment shortly when I have some more time. It is possible to start with 4BVH for one simulation since it would be easiest to set up, but that would not be enough in itself.
With the loop replacement, I do not have information on side chain RMSDs in the loop environment; that may require some structure prep as well. Hence the importance of the structure prep tasks, perhaps while running the sim on 4BVH.
The issue is that I need both open and closed loop simulation / comparison. I will provide more details.
Considerations in the choice of structures for use in the coproduct simulations:
--binding energies with open,closed conformations of loop required
--consider our use of 4FVT w/ loop replacement from 4BVG for INT loop conformation.
4FVT was positioned or intermediate/NAM were prepared according to protocol provided by Ping.
we also did simulations starting from intermediate complex xtal structure with loop replacement from 4FVT,
but these are not being reported in this paper
--similarly for coproduct, loop replacement is required
-preparing as in case of 4FVT with loop replacement from 4BVG (with coproduct ligand) would require similar structure
preparation for latter
-could alternatively start from apoenzyme xtal structure and replace with a closed coproduct binding loop; use of 4FVT and 4BVG (INT) loop would require less structure preparation since that already done by Ping
-alternatively, starting from coproduct complex xtal structure, one could replace loop with apo loop conformation (open). But this would be more analogous to the use of 4BVG with
loop replacement from 4FVT, which we are not reporting in this paper.
-using different open and closed loops would require us to report all the relevant conformational energies in the table.
We should bear the above in mind and make decision after you find the structure preparation procotol and are ready to run / comfortable with running the NAMD simulations.
RSK:Attached a figure to illustrate it.
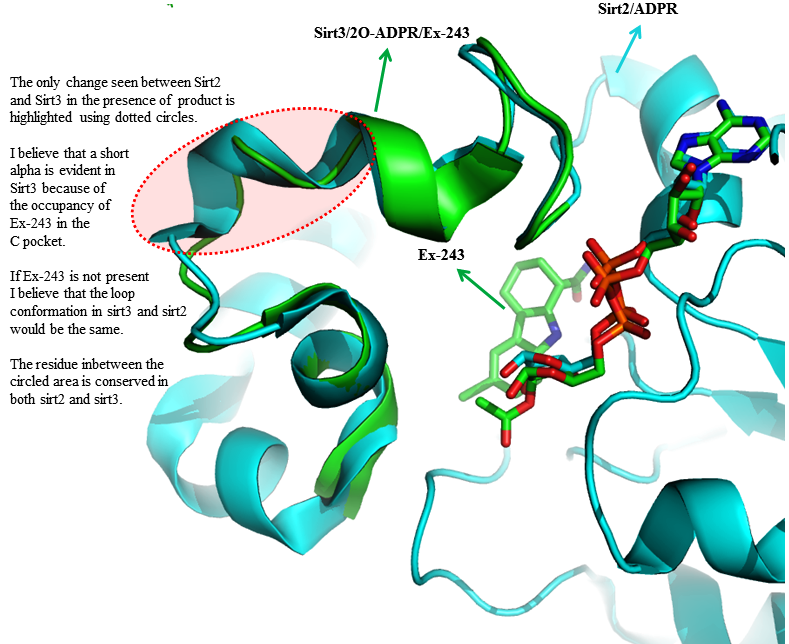
RSK: I think that it would be straight forward approach, and we many not have to include loop and side chain modelling in our protocol for generating a Sirt3/product complex.
RSK: If you give the go-ahead then I can start learning how to set up an MD simulation in NAMD and try using the same MD parameters which Ping had used for the other simulations.
RC: It is relatively straightforward to learn how to set up the MD simulations in NAMD given that you have all the command history and input files. If you look at the input files generated by Ping I believe you will
see similarities with Amber. You should be reviewing this concurrently.
RSK: Okay, I will look into the reports prepared by Ping. Once, I have the structure preparation protocol ready, I will send it to you for your approval before I go ahead with the computation work.
RC: I would like to see updates on what you are finding in terms of the structure preparation protocol to verify you are on the right track,
given that Ping and I worked on this. E.g., there was no loop prediction in these protocols so you do not need to look into those notes for the purpose of this structure preparation.
BTW, have you found the plop code?
--Regarding the tools used for structure preparation:
-note that we have plop source code and executables installed on the cluster (probably slave004). I have mentioned this to Ping before but he never needed to use it and preferred to use the prime gui. The path is probably listed somewhere on the wiki. I believe it is in a protein subdirectory of the ~rajchak/Simulation directory.Some of the protein design python scripts described on the software dev page of wiki used it. This is essentially the academic version of prime. It can do side chain optimizations, minimizations for which we used prime in our structure preparation protocols but you need to look into the input "con" files and look up the commands.
RSK: Thanks, I will look in to it. A side not here, I believe that Prime has more advanced functionality especially in the case of loop modelling using some advanced implicit solvent models ( not through GUI but command based interface), touted as the Frisner loop modelling protocol. I once came across a log file that hinted me that Ping had even attempted that protocol.
RC: Yes he did use the long loop refinement protocol for some work (specifically, filling in the missing loop segments in Sir2, and some attempts at SIRT3 loop prediction, which were to be continued later, not in the immediate papers). He also identified the input file commands corresponding to the sampling algorithm in that case. You and I discussed this topic as part of questionnaire. It's true that it is not part of the plop source we have, which dates back to 2005. But, we're not doing loop prediction in this paper and it wasn't part of his structure prep method. Only side chain optimization was used. There as well, though, there may be some differences since the side chain sampling algorithms probably changed somewhat. But in any case, it's useful to know how to use this code.
Anyways, I am not confident yet to comment which protocol he setlled finally. But, looking through all files and report, I believe that we could figure it.
RC: Please note that for side chain prediction step, there was also a validation phase of the study (prediction in native environment) reported.
-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
-regarding schrodinger software, please check with Sherry on when the license will start and also ask her whether there is prospect for obtaining it earlier. But in any case we will not proceed until the protocol is clearly established.
we would then decide whether to use plop in the interim depending on the time required to use it and the fact that we would repeat using the latest version of prime later anyway.
-indicate when you are comfortable with using the required NAMD tools.
RSK: I agree that we should wait until we figure out the protocol for "Loop modelling" . I did briefly talk with Sherry regarding this, and she informed me that the start date for our licence will be June. Once we figure out the protocol, may be we can request them for a one week trail version, so that we could get the job completed before June.
RC: Ok
--I am working on the SI of the paper and will be posting a draft soon. I would like you to look at some of the missing computational sections
and comment on whether we want to add any more existing figures to this section.
I will use the pmc-at patents dropbox folder to share these docs since that is apparently where other shared paper drafts are located.
There may be other upcoming paper related tasks other than coproduct simulation as well.
I have posted two SI docs:
-Supporting Info short 2-26
-SI computational data
in the pmc-at patents dropbox folder.
Please see pg. 11 of the former; this computational data section has not been updated since an early draft.
The required data will mostly come from the latter doc. I have edited the latter to highlight the most important figures and tables
and reordered them. The ones listed as "optional" should not be transferred at this time.
Please transfer the others to the computational data section and note the missing figures (Sir2, e.g.)
Please indicate whether you have figs that can be used here or whether we should reconsider this. Here, our main goal
was to show our conclusions were not limited to only SIRT3, since the paper is not on SIRT3.
Regarding the 4FVT/4BVG xtal structure alignment fig that is currently in paper body, I believe we will be moving that to SI,
so you can include a copy of that there as well.
Also, please indicate if there were any other figs that you had prepared that we were considering putting in SI that are not currently there.
(E.g., regarding coproduct complex, if you have a structure of that for a Sir2, we may consider including it in an alignment, though this is not a priority.)
See also the requested change to the xtal RMSD plot to add the coproduct RMSD.
RSK: I have placed revised draft in the Dropbox. In case if you find that it is not shared with you please let me know , i will share it again.
RSK: The missing figures are Sirt2 and a 2D plot showing the non-bonded interactions of the Intermediate product with Sirt3
I may also ask you at some point to revise some non-computational figures for the paper using appropriate image editing tools.
You may want to ask Alok/Guan about the tools they used for these and possibly install them on your desktop.
RSK: sure.
Regarding methods (which are mentioned in one SI section), most of those will be handled last. For now, identifying the structure preparation methods (above) are the priority.
RSK: Okay will look into it and revise it appropriately as per the comments.
Another pending task that is related to the last table in this doc is the time series plots of MM-GB(PB)SA energies. I believe you mentioned
the scripts were ready to generate these plots. I don't believe we followed up yet on those scripts or plots. Please prepare them when ready, and insert those time series
plots in paper body.
RSK: Yes, the scripts required for automation are ready, but we got side tracked form this. I will set up the run today, so that it will run tonight and i can get working on the plot tomorrow morning.
RSK: Will also document the script and place it under the Software development header on the wiki, for further use
We had communicated about the associated pdf plots, which had been assigned earlier to Ping; those can be inserted in SI after the above are complete.
RSK: Okay, to extract these energies , I will have to write another script that fetches the energy of each frame. I can get that done.The script is not ready, but I can get it working.
RC: This can be done after most of the above are complete (not necessarily including actual implementation of the coproduct structure preparation protocol).
4/27/2016
RSK: I did a page check using PNAS article sizing tool. The generated pdf document is attached below. Although all the required references have been added, I will have to recheck the numbering of the references once more.
This is just to provide you an idea on the page length the current draft.
pagecheck.pdf
RSK: Revised version incorporating your latest comments.
pagecheck1.pdf
RC:
-- You didn't insert the double reciprocal plots (linear plots with eqn insets); as mentioned earlier. these are not experimental data. The experimental plots should not be included.
-- The comment about size of rmsd plot vs size of structural fig applies also Fig 6. The latter structural fig is important because it is based on sim data and is not clearly readable.
-- Please also upload the latest doc version (or you can store them in dropbox if you prefer).
5/2/2016
RSK: Please find attached a revised version of the PNAS manuscript in PNAS style ( page length check) . This version includes the double reciprocal plot.
pagecheck2.pdf
RSK: Also let me know if the size of figure 6 is okay . If not I can redo it by reducing the size of the RMSD plot shown in panel A. In the revised figure I decreased the size of panel A and enlarged the structure figure in panel B.
RSK: A word document of the working draft of the PNAS manuscript is also attached below. Note the figures are embedded within the draft for ease of understanding.
PNAS_draft_2May2016.docx
RC: Yes, the size of the RMSD plot may be further reduced in that fig. Or, it may be advisable to combine Figs 4 and 5 (since these are xtal structure figs rather than our results) and separate 6 into two parts so the structure from
our simulations is easier to inspect.
You can see what works best, not a major priority.
RSK: Okay, will try both the options.
4/22/2016
RSK: A working draft of the manuscript is attached below. I need to add a reference which you had indicated earlier.Although, I have added the figures to the draft it is still a work in progress.
PNAS_draft_22Apr2016.docx
4/22/2016
RSK: A revised RMSD table is added. Please consider it as the final RMSD table as includes values for all the compassion we had made. It includes RMSD for the
a) NADP part
b) Peptide part
c) NAM (Applicable in one case only --Sirt3/Int/NAM with 4FVT loop vs Sirt3/Int/NAM with 4BVG loop)
RMSD table.docx
.
RSK: Since all the supplementary data work is complete, I will start working on revising the main part of the PNAS manuscript draft based on your latest comments
RC: Since the methods for RMSD calculation have been worked out, would you also be able to provide RMSDs for substrates for 4FVT compared to 4BVG (to
compare substrate conformations before/after NAM cleavage)?
RSK: I think that I have reported the RMSD values for particular pair in the file "RMSD table.dock" which I uploaded today morning. It should be the first pair listed in the table. In case that is different from what you had asked for, please let me know . I can get it done.
RC: Ok got it.
RC: I have been reviewing the Task list 2 and updated manuscript.
A few immediate comments:
a) why is one of the RMSD plots apparently repeated twice in the manuscript draft?
b) the kinetic schematics (double reciprocal plots not based on experimental data, but rather schematics with equations inset) in the biorxiv manuscript are to be kept in the revision, since they are not experimental data. Please put them back in.
--These schematics are not in the current draft above; the ones I mean are the double reciprocal plots with inset equations.
c) Regarding the apo (3GLS) structure and its cofactor binding loop conformation/flexibility, please note that some prior literature has discussed open and closed conformations of this loop. See in particular the PNAS paper from Steegborn on Ex-527 mechanism (sirtuin inhibitor). This paper seems to claim that the apo conformation of the loop is "open" and that NAD+ binding stabilizes a "closed" conformation, followed by NAM cleavage resulting in an even more "closed" conformation. We are familiar with the latter conformational change, but it appears that your analysis suggests that for SIRT3, apo and ternary (NAD+ bound) loop conformations are quite similar. You may check whether some of these comments were based on Sir2 structures.You may also refer/compare to Ping's ppt on loop conformations in this regard.In any case, comments need to be made here regarding consistency with literature.
RSK: I think that the definition of open/closed loop conformation is more closely tied to the position of Phe 157 residue with respect to the ribosyl ring of ADPR than the overall loop conformation. I presume that the paper from Steegborn group ( PNAS describing EX-527 mechanism of inhibition) which introduces the terminology called open/closed loop conformation in loosely based on this . Based on my understanding of the literature it sounds that when Phe 157 is stacked face-to-face against the ribosyl ring of ADPR , it acts as a hydrophobic shield that blocks the reaction intermediate form participating in base exchange reaction with nicotineamide. Historically, these findings were described for Sirt2 and I think that extrapolated this information as a similar mechanism is evident for Sirt3.
RSK: I have attached a figure that dillustares this.
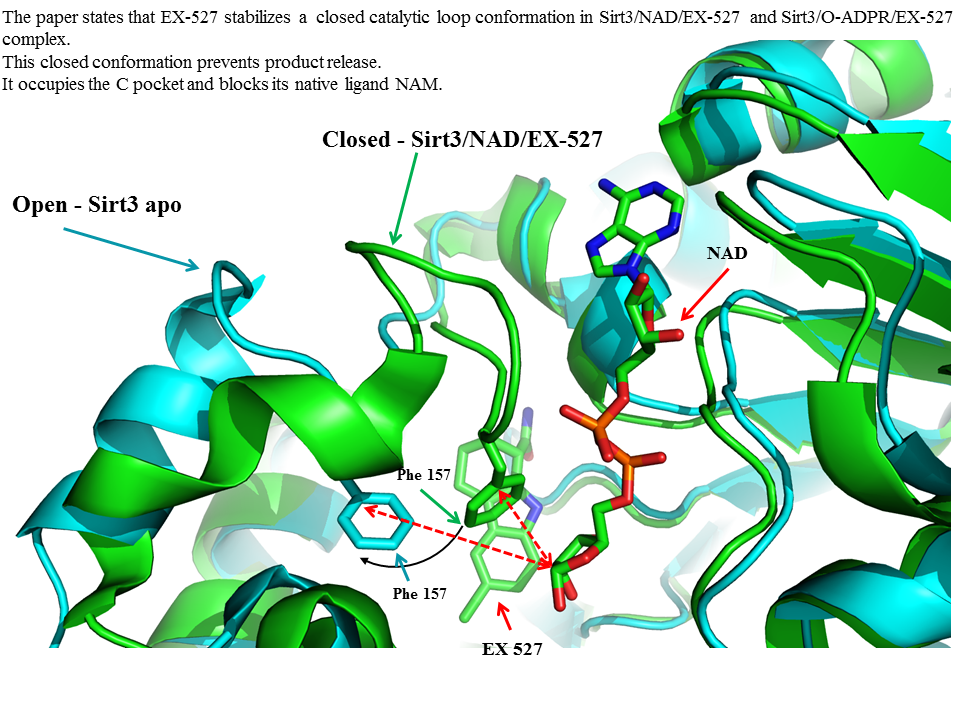
RSK. I also have a general comment regarding a statement in their PNAS paper " Comparison of our Ex-243 complexes with apo Sirt3 (PDB ID 3GLS) (13) and Sir2Tm/Sirt3 structures with occupied NAD+ pockets [Sir2Tm/NAD+/p53 peptide, 2H4F (16) peptide, 2H4F:--- indeed shows that Ex-243 stabilizes the loop’s closed conformation also adopted on NAD+ binding (Fig. 4B; Fig. S4)" . This statement raises two questions
a) The PDB id 2H4F which they does mention doesn't have the co-factor loop region resolved.
b) They allude to Fig 4B and S4 in their above statement. However Fig 4 is an comparison of apo Sirt3 (3GLS) vs Sirt3/O-Adpr/Ex-243 and not Sirt3/O-Adpr/Ex-243 vs Sirt3/NAD+/peptide. Also, I dint see any figure S4 in the supplementary information as stated.The supplementary information ends with S3.
RC: Ok. I provide some comments on the issue of NAD+ vs coproduct complex loop conformations ("closed" vs "open") below.
RSK: Regarding your question as to why the the RMSD plot shows that apo Sirt3 (3GLS) and Sirt3 /Ternary complex (4FVT) loop conformation are quite similar? I did check the structures once again by superimposing them. The data for the plot which in fact was generated by Ping and re plotted by me sounds to be right. I think globally the loop conformation tend to somewhat similar, but the position of the Phe 157 is strikingly different.I think the PNAS paper by Steegborn is basing their definition of open/close based on the position of Phe.Attached below is a figure to illustrate the correctness of the RMSD plot.
RC: Yes, our by-residue RMSD plots also suggest the RMSD between the apo and ternary complex loops is significant around Phe157.However, it seems that what they call the "closed" conformation in presence of NAD+ is much more open than the "closed" conformationin the presence of intermediate or coproduct.
RSK: Your observation is right. The Sirt3/2-OAcADPr/Ex-243 complex is relatively more closed than Sirt3/NAD+/Ex-243 and Sirt3/ADPr/Ex-243 complex. I see that the short helix rotates inward towards the active site.RSK: The conformation of the co-factor loop is largely similar in Sirt3/NAD+/Ex-243 and Sirt3/ADPr/Ex-243 complex.
-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
RC: I need some more information on this topic.
-- in the latter picture above you show NAD+ along with the apo and ternary crystal structures of the receptor. That is fine.-- in the former picture above, you mention the closed loop conformation and annotate that as SIRT3/NAD/Ex-527. Please confirm that the receptorwas obtained from the xtal structure for that complex. The paper mentions that NAD resides in nonproductive (perhaps AB) pocket conformationin presence of Ex-243/527. The loop conformation looks quite different from that in 4FVT.-- please show for comparison the xtal structure for the coproduct complex with Ex-527. If you can show all of these in representations similar/aligned to those in the paper draft,it would be convenient (though not essential).-- you may also superimpose the xtal structure, if available, of the coproduct complex (no Ex-527) with that of the intermediate complex (4BVG).I would like to compare the loop conformations and determine whether the loop "closes further" upon conversion of intermediate to coproduct. -- finally, please add the coproduct loop conformation (no Ex-527) to the by-residue RMSD plot in the paper from xtal structures, since this complex is important.
If you don't have the coproduct xtal structure, you can use the Ex-527 coproduct complex for this purpose.
RSK: Please find attached a report in response to the above questions. I will be commenting/completing the other tasks which you had classified to be of lesser priority shortly.
Loopreport_may4th.docx
RC: Comments on the report:
-please comment further on the consistency of a) your latest statement that apo loop conformation is more disordered/flexible than ternary loop conformation
b) our earlier discussions where I indicated Ping may have stated that apo loop conformation is more flexible, but you indicated this is not the case, along with supporting information.
This is important to our presentation. Was a) based on SIRT3 or other sirtuins?
RSK: Response for question A & B ( I am copying this email transcript for documentation purpose. This has already been read and commented by you )
I realize that my latest comment pertaining to the cofactor loop contained in the report sounds self-contradictory.
The reason for the confusion stems from the difference between my understating of the literature ( mostly review articles) statements and individual Sirt3 structural observation.
Most literatures (review articles in particular) carry an generic statement that the cofactor loop is highly disordered in apo sirtuins which becomes ordered upon substrate/cofactor binding. I believe that this particular observation that was initially reported for yeast Hst2 eventually became more of an generic statement for all “SIRTUINS”.
I did pull up some structures of yHst2 in pymol, and I did see that the loop is partially unresolved in apo form but not for structures complexed with intermediate and the reaction product. However, I dint look more closely at them by plotting the B factors in the interest of time.
I believe this to be reason for the confusion in my statement and I convinced that our earlier observation on Sirt3 co-factor still holds good.
When , I find time I will look more closely in to the behavior of sirt2 vs Sirt3 , to see if they are specific to certain sirtuins
RC-in the first figure one structure is annotated ADPr Ex-243 complex and the comments below refer to intermediate. Did the paper report a complex of Ex-243 with the intermediate?
Is this really the intermediate complex loop conformation?
RSK: I am sorry it should have been commented as the "Substrate Complex" and not as "intermediate complex".The error has been fixed and I have upload a revised version of the draft. The annotation of the figure is right.
Secondly, the paper does report three intermediate structures namely
4BVE (Sirt3 thio-intermediate)
4BVF (Sirt3 thio-intermediate)
4BVG (Sirt3 native intermediate)
All these complex do not contain the Ex-243.
-as time permits please provide a representation that highlights the interactions of loop with intermediate (not just Phe). This could include representation of some of the side chains.
Is it primarily the alpha turn that is involved in these interactions?
RSK: Attached is an figure illustrating the interactions of the "reaction intermediate" with the co-factor loop.
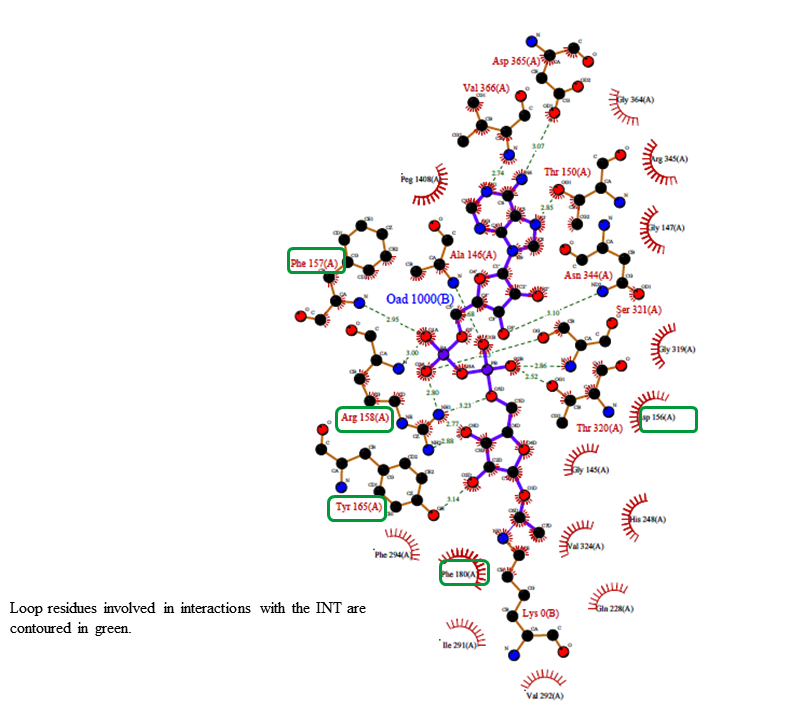
RC: I assume this is for the INT (4BVG) loop conformation (after MD). We should generate a similar ligand interaction diagram for the open loop conformation (4FVT conformation from our simulations).
-yes we are doing detailed studies on loop conformational changes that occur upon formation of intermediate complex with attention to its role in catalysis
-for future reference and organization, the report should be combined, filed together with, or cross-reference Ping's loop report/ppt, which I believe also contains other Sir2 structures.RSK: I will merge this loop report with Pings report on loop modelling.
-priority is still the binding affinity calculations below and other paper related tasks.
RSK: Binding affinity calculations completed.
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- related, there is the issue of whether/to what extent the "closed" conformations of the loop stabilize coproduct binding, as discussed in the Ex-527 paper.
Please comment on what would be the easiest calculation for you to do in this regard. You could do MM-GB(PB)SA binding affinity calc (as we did for NAM)
with the ADPR-Intermediate as a ligand (perhaps turning off interactions of the peptide part) or you could do the calculation with the coproduct as ligand.
The former could probably be done by rescoring existing trajectories whereas the latter may need to be done from scratch, please look into it.
RSK: You are right, we can extract the MM/GBSA and MM/PBSA binding affinities by rescoring the existing MD trajectories by defining the ADPR-intermediate as the ligand. This would be the most straight forward method. However, I am bit skeptical about turning off the interaction from the peptide part of the ADPR. Firstly, I am not sure if we can define a substructure of a molecule (ADPr) as a ligand ( excluding the peptide that is covalently linked to the ADPr). Secondly, excluding the peptide part of ADPr-peptide intermediate from the MM/PBSA and MM/GBSA may not mimic the actual coproduct ADP-ribose, because the presence of the peptide part in the simulation could have influenced the dynamics and the conformation of sirt3 and the ADPR moiety itself.
RSK: I did perform an MM/PBSA and MM/GBSA calculation on the existing trajectories by defining the "reaction intermediate" as the "Ligand". The values obtained are tabulated in the attached document.RSK: MM/PBSA and MM/GBSA calculations reveal that the intermediate loop conformation (as seen in 4BVG) stabilizes the "reaction intermediate" over a ternary loop conformation (as seen in 4FVT). To put it the other way around in terms of structure, we could say that a closing of the co-factor loop stabilizes the ADPr intermediate.
MM_PB_GBSA_INT_May5th.docx
----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
RSK: However for calculation with coproduct (ADP-ribose) we may have to do an MD simulation.
RC: You are right, on subsequent consideration I also concluded that we should score the intermediate as is, not as a substructure, and do the coproduct simulation from scratch.
That quick fix is not warranted.
Both with the intermediate loop conformation since that is what we are studying in the paper.
I would like to add some such calculation to the table in the paper.You may start with scoring the existing trajectories with ADPR-Intermediate since those will be useful to have anyway, and then (or perhaps concurrently) move on to coproduct.
RSK: Okay, I will begin by scoring the existing trajectories for extracting the binding affinities of the ADPR intermediate. Simultaneously, I will try setting up a MD simulation for the coproduct also.This would require some time as we need to model the system first and then employ same input MD parameters employed by Ping for the Sirt3/ADPR intermediate. Moreover, the simulations have been carried out in NAMD and not Amber. So, I need to transition form AMBER to NAMD. I dont foresee that transition to be hard for me because the fundamental steps in all MD packages are similar, except for the commands and flags.
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
I believe I may have asked Ping about this claim of the paper before, perhaps you can have a look at old correspondence or postings. He seemed to be somewhat skeptical of
some claims in the paper, as I recall.
RSK: I will comment on it after looking through all previous comments and communications. Any idea as to what specif claim in the paper is contentious so that i can look more closely?
-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
Looking at the PNAS paper figures (and the text), it appears there may be direct contacts between Ex-527 and the coproduct/NAD (latter in nonproductive conformation). It appears Phe157may interact with Ex-527 and Ex-527 in turns interacts with the coproduct/NAD. It is not clear whether there are direct contacts between Phe157 and coproduct/NAD. Ex-527 I believe is in the C pocket,not Phe157, and Phe157 may also rotate. It is also not clear whether it is Ex-527 or Phe157 that is pushing nicotinamide out of the C pocket in the NAD:Ex-527 complex. Of course we can explore this in more detail later, but you can provide your comments since you have access to the structures.
RSK: You are absolutely right. Ex-243 engages in an stacking interaction with NAD and Phe 157. Ex-243 occupies the C pocket which in turns pushes the nicotinamide moiety of NAD away form the C pocket to the Acetyl Lysine channel resulting in non-productive conformation. Phe 157 which used to be positioned above the B pocket in a productive conformation (sirt3/NAD) is now recruited into the C pocket in the presence of Ex-243 (non -productive conformation).Attached below is an image that illustrates these non-bonded interactions involving Phe 157 and NAD with Ex-243 ( also see the figure provided in the Loopreport_may4th.dock for more details)
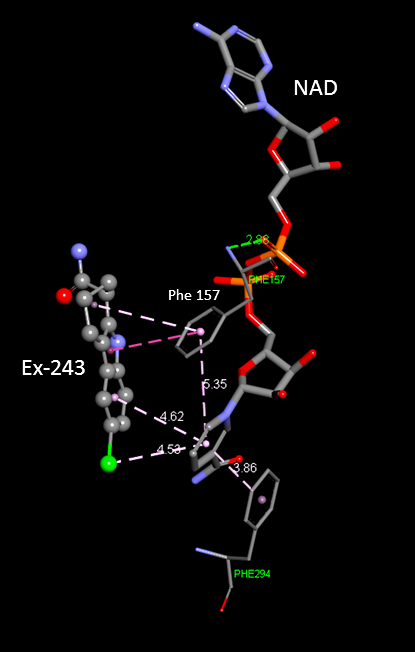
RSK: I believe that inhibitor Ex-243 push the nicotinamide away form the C pocket to the Acetyl-Lysine channel resulting in a non productive conformation for the following reasons.
a) Ex-243 seems to be an structural mimetic of NAM and precisely form the same interaction as NAM b) Free NAM when complexed with Sirt3/NADP also results in a non-productive conformation. c) Binding of Ex-243 recruits the Phe 157 to the C pocket.
-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
--Also, please remind me of our previously reported (e.g., in PLOS) values of NAD+ binding affinity in the AC vs AB pockets (latter in presence of NAM in C pocket).
RSK: The MM-PB(GB)SA binding affinity estimates for NAD+ reported in the PLOS paper for the AC pocket ( productive mode ) is -23.19 kcal/mol and -16.15 kcal/mol for the AB pocket ( non-productive mode).
Please treat this as a priority.
RSK: Okay I will start working on it.----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
d) regarding the preparation of the starting structures for MD simulations (esp those with loop substitution), do you have access to a summary of methods used for this structure preparation among the documents provided to you from Ping? This will be relevant to subsequent Methods work.
e) The RMSD plot that accompanies one of the structures in the same figure might be made an inset so as to increase the size of the structure and decrease the size of the RMSD plot.
4/21/2016
RSK: Responses to your latest comments ( regarding the supplementary data) are addressed in the draft attached below.
Task 2.docx
RSK: A cleaned up version of the supplementary information is attached below.
SI.docx
RC: Regarding your comments from task 2 doc:
- your understanding regarding free energies is correct. Ping was omitting the entropy contribution in the tables
he generated for papers/reports. Ping didn't use \Delta G notation rigorously, he reported approximations.
As noted our next papers (following this one) will be more accurate in various respects.
- yes we can consider providing a pdf of the type you mentioned.
we will discuss this and decide on format of presentation after seeing the trajectory figures requested.
You will notice that I had asked Ping to provide this pdf for various simulation times in one of his task lists.
He didn't have time to prepare them. I am not sure if that was included in your task list.
4/12/2016
RSK: I have finished the tasks relevant to the MANUSCRIPT draft which was placed in the Dropbox.
Few miscellaneous task required for the supplementary information are pending, and I am working on it and I hope to get it done by Wednesday.
RC: a) re-introduce the missing figures from the current paper draft as per their positions in biorxiv (everything -except- experimental data figures).
RSK: This task is complete. I have also incorporated the general scheme for Sirtuins deacylation from the biorxiv manuscript. All other plot relevant to experimental data are omitted.
Track changes option has been used.
Manuscript1.docx
RC: b) prepare a working file (appendix/SI) which includes all the latest figures not included in paper draft. We will organize this later.
RSK: I hope to upload the completed Supplementary Section by Wednesday
Pending task ( Generating more data points from the MMPBSA and GBSA calculation) is the pending task
I expect this calculation to take more time. Looking at the log file I see that each PB calculation carried out by Ping has taken 8-9 hrs. This data will be needed for generating more data points for Fig 7.
RC: d) add the new reference; check there are no missing refs
RSK: Two new references have been added and the reference numbers has been edited appropriately
RC : e) do a PNAS length check (use their online tool; assume extended online format)
RSK: I did a page check using their online tool.
Two issues I did notice a) The significance statement exceeds by ~ 10 words (slightly off the mark)
b) even without the computational method section it runs to 9 pages. (computational methods can be carried over to the supplementary information)
Note: PNAS also allows a 10 page submission and 6 page submission
RC: f) The table on MM-GBSA energies should not highlight the binding affinities only, since the free energies of loop conformational changes are
relevant. You may add a column or otherwise rework the table and its title to reflect this and report \Delta G for loop conformational change as well
(in this sense it departs from previous tables)
RSK: The table has been revised appropriately. I have also added a short paragraph (4 lines).
RSK: The Plot of MM/PBSA and MM/GBSA energies vs time has been revised (Fig 7).
RC : Task G
RSK: This task is relevant to Supplementary information. So, these changes are not reflected in the Main part of the manuscript. I will comment on along with the supplementary materials on Wednesday.
RC: h) Color coding of structures in all figures must be consistent
RSK: The color coding are consistent, I did check it.
RC: i) Make a plot of B factors from the MD simulations of non-crystallographic complexes (e.g., INT:NAM complexes)
RSK: This task is relevant to Supplementary information. I will upload it with supplementary information by Wed evening
RSK: The purpose of the draft is help whittle down the pages of the original version of Task list 1 ( which i uploaded yesterday ) for ease of reviewing.
Cleaned_version_Task1.docx
RC (4/12): I have placed a paper draft ("biorxiv_manuscript_4-12_eqns_inline_no_figs") with working versions of the computational figures in dropbox PMC-AT patents (which is the folder in which we were sharing docs relevant to this paper). I am inviting you to that dropbox.
Therein you will find previous drafts used for biorxiv including figures. However, these earlier figures were removed from the latest paper draft. We would like to get an idea of length.
In addition to completing your pending tasks, you need to:
a) re-introduce the missing figures from the current paper draft as per their positions in biorxiv (everything -except- experimental data figures).
b) prepare a working file (appendix/SI) which includes all the latest figures not included in paper draft. We will organize this later.
c) clean up/merge the inserted computational figures as needed
d) add the new reference; check there are no missing refs
e) do a PNAS length check (use their online tool; assume extended online format)
f) The table on MM-GBSA energies should not highlight the binding affinities only, since the free energies of loop conformational changes are
relevant. You may add a column or otherwise rework the table and its title to reflect this and report \Delta G for loop conformational change as well
(in this sense it departs from previous tables).
g) Regarding the figure
"Fig ---- : Superposition of the time averaged MD structures of Sirt3/Peptide/NAD ternary complex (4FVT – orange) and Sirt3/Intermediate complex (4BVG – green). Differences in the conformations of the co-factor binding loop and the position of the Phe residue are highlighted. Individual subsites are highlighted."
some follow up work is needed:
- please add corresponding entries to the global/loop RMSD table and/or corresponding per-residue RMSD plots for these simulations with respect to xtal structures
- comparing to the B factor plots, comment on which loop segment displays the greatest flexibility vis-a-vis the structural motifs like helix,
alpha turn.
- Generate a figure analogous to "Time averaged MD structure of Sirt3: INT: NAM modeled using the 4BVG loop (Green) superimposed onto 4BVG Xtal structure (Global RMSD = 1 Å)",
but for the apoprotein MD average and the 3GLS xtal structure instead. Related to the comment in the draft about verification that the apo loop conformation is similar to that in the ternary complex, verify that despite this conformational similarity in the xtal
structures, the loop in ternary complex is more flexible (compared to the flexibility in both the INT and apo complexes) and that this apparent in the RMSDs of the MD averages to the respective xtal structures.
- add entry for the apoprotein MD average with respect to 3GLS to the global/loop RMSD table
h) Color coding of structures in all figures must be consistent
i) Make a plot of B factors from the MD simulations of non-crystallographic complexes (e.g., INT:NAM complexes)
Note that in the current working draft, we are assuming there will be no experimental data included.
SI will be further edited after you complete these tasks, followed by Methods.
RSK(4/12) : Okay will go through the draft (biorxiv_manuscript_4-12_eqns_inline_no_figs.dock) and carry out the required changes
Regarding the latest task list revisions:
- I need more info regarding the RMSD issue. Specifically, which structures are you using to compute RMSDs?
Do you mean the problem is occurring when computing RMSDs for ADPR moiety in intermediate with respect to ADPR
moiety in NAD+? Or even ADPR-INT to ADPR-INT?
This is potentially important to the analysis since I would like to see how the conformation of the intermediate compares to that
of substrates, esp given the significant impact the reaction has on the optimal loop conformation.
RSK(4/12): RMSD compassion is just an technical issue, which could be fixed, Its not specif to any chemical moiety. The only issue is that it involves manual/visual intervention and requires time. Since, you mentioned that it is important, I will get it done.
To compare RMSD for small molecules the following conditions need to be satisfied.
A) The "reference " and the "target" structure should have the same number of atoms (ie, you can compare NAD+ conformer A and NAD+ conformer B) / or you can compare a selected list of matching atoms between conformers.
B) The atoms in the target and reference structure must have the same naming convention ( ie, if the amino nitrogen in the adenine ring of ADPR is named as N1 in the reference structure then the corresponding target structure should also have the same atom name N1 or not any other naming convention (N, N' etc)
C) The atoms listed in the target file and reference file should be in the same sequential order.
Basically, I need to ensure these conditions are satisfied before computing RMSD
- For the MD trajectories figure, in each case I would like to use the MM-GBSA energies of the complex on
which the simulation was actually run (not binding affinity), and I would like to report data points at higher frequency --
assuming you have access to this data.
RSK(4.12): I don't have the other data points, but I will try to collect more data points from the trajectory.
Firstly, I will try to reproduce the MM/PBSA numbers reported by Ping by rerunning the trajectory for one complex.
If, I can reproduce the numbers then I will go ahead and collect more data points form the trajectory.
I will also have to locate the mmpbsa and mmgbsa input files to ensure all the parameters used for the calculations are consistent to reproduce it.
Will let you know how it goes.
Note as mentioned earlier that the computational work would be done/reported more rigorously in the subsequent longer papers,
for which drafts are under development.
4/11/2016
RSK: Attached below is the latest revisions for Task 1.
RSK: The latest comments form your end is embedded into the documented dated (4/3/2016)
RSK: Since the document carries over all the revision, I have highlighted your latest comments in yellow, so that it would be essay to track down the revisions. ( If needed I can prepare a cleaned up version, since all the listed items in Task I is now complete)
RSK: My comments and revisions are dated 4/8/2016.
COMMENT/Issue: One issue that popped up during the course of revision.
a) Going through the document prepared by Plin, I see that he mentions carrying out simulations starting form 4BVG-INT-NA with native loop and 4BVG-INT-NAM with the loop residues grafted form 4FVT.
Although I was able to locate the former trajectory, the latter trajectory is not available in the GPU node.
I will also search for the presence of the said trajectory in the other nodes of the cluster.
b) Another trivial issue: you had also asked to measure the RMSD for ADP and INT moiety also. However, there is an technical glitch in computing RMSD for small molecules. I have explained it in detail in the document.
However, this issue could be overcome, but it needs manual intervention and time.
Apart form these two issues all other listed comments have been duly addressed in the revised draft.
Vijayan_task_list1_4-11-2016.docx
4/4/2206
RSK: Will work on your comments. If time permits, I will also go through the previous system admin task listed on the wiki, which you had asked me to familiarize with.
4/1/2016
RSK: Attached below is a document that contain revisions for Task 1.
The latest comments from your end is dated RC 3/22/2016
and my comments and revisions are under RSK (4/1/2016):
This documents also contains all prevision revisions.
RC: Thanks, I will review. First, please let me know if you have completely finished all listed tasks (and cannot proceed further without
feedback) or whether you are still working on some of them.
RSK: There is only one listed task which is pending your feedback and incomplete. I was not able to complete the RMSD plot task for the loop region of Sir2TM similar to the one we have for Sirt3 using information form the Crystal structure.
The reason which I have commented in the document is copied below
"Regarding your question for preparing a similar figure for Sirt2Tm, I foresee an issue here. I happened to look through all structures of Human Sirt2 and Sir2TM available in PDB. It appears that all Sirt2 (ternary and intermediate) complexes have the cofactor binding loop unresolved (missing density)."
RC (4/3): Yes, we were aware of this (see Pings report); can't create structure alignment for Sir2 without the missing residues? Not a priority.
Also, another less priority task which was not meant for paper but for analysis stated " RC (319): In addition, separately for the purpose of analysis (not publication), please provide the following RMSD plots (all for loops only):"
a) 4FVT/4BVG (pdb) – 4FVT:INT:NAM (md average)
b) 4BVG/4FVT (pdb) – 4FVT:INT:NAM with loop replacement from 4BVG (md average)
c) 4FVT/4BVG (pdb) – 4BVG:INT:NAM with loop replacement from 4FVT (md average)
d) 4BVG/4FVT (pdb) – 4FVT:INT:NAM (md average) " is to be completed.
Currently, I am working on it and it may take a day for me to get it done.
Rest all the listed tasks are complete.
RC (4/3): In addition to loop RMSD add ADPR moiety (including in NAD+) and peptide moiety (including in INT) RMSDs to the RMSD table at end of doc that is currently underpreparation
Vijayan_task_list1_4-1-2016.docx
RC (4/3): It is good to see that many of the issues with MD average structures were resolved,
including a demonstration that the conversion of NAD+/peptide to NAM/ADPR-intermediate resulted in significant conformational
changes for the loop modeled based on 4FVT (consistent with expectations).
In the interests of time, I am providing comments directly here regarding your assignments for the week; please incorporate them with appropriate date -- RC (4/3) -- within the working document as
appropriate:
- - Regarding B factors of apo loop, a comment: flexibility of apo complex mentioned by Ping might have been referring to other parts of the protein like the peptide binding pocket,not the cofactor binding loop. In any case, indicate specifically how long the apo (3GLS) simulation was run, given that the B factors seem quite a bit lower than those of the 4FVT conformation, which appears to be a similar conformation
- In all captions indicate what complexes the PDB entries correspond to
- Regarding annotation of residues in structure alignments: for understanding purposes you might highlight just Arg 34 (Sir2 numbering - use appropriate SIRT3 numbering) in the MD simulation figs
- In new MD average superposition, please specify which substrates are which (e.g., which NAM is which -- there are two NAMs shown).
- Prepare an analogous MD average fig from the simulation data starting from 4BVG rather than 4FVT (not necessarily for publication)
- Make plots of time series of MM-GBSA energies for the MD trajectories based on following caption draft:
Figure. Molecular dynamics trajectories for SIRT3 complexes after structure preparation. Time course of MD trajectories: MM-GB(PB)SA energy vs time
- A) SIRT3/INT/NAM prepared from 4FVT w/ loop (res 155-178) replacement from 4FVT; B)SIRT3/INT/NAM prepared from 4FVT
| Complex |
Global heavy atom RMSD |
Table, include a new column showing cofactor binding loop RMSD
- In "Fig ---- : Superposition of the time averaged MD structures of Sirt3/Peptide/NAD ternary complex (4FVT – orange) and Sirt3/Intermediate complex (4BVG – green).
Differences in the conformations of the co-factor binding loop and the position of the Phe residue are highlighted. Individual subsites are highlighted."
you mention a helix is forming in the INT complex simulation, but in the subsequent time-averaged MD figure of INT:NAM complex, it appears this is not the case.
Please comment/clarify and also provide the global RMSD of the former (latter was already provided).
- add the reference on ELT inhibitors of SIRT3 (3JSR) to the end of the doc for citation in paper
- Perhaps in an appendix or a separate document, provide figures depicting the starting structures for each of the simulations for which MD average structures were provided in this report
RSK: Attached below is a document that contains my brief responses and clarifications to the comments which you had addressed to me during the course of revision.
I see that the MD average structures needs to be replaced and I realize that is my immediate priority.
Also, please note that I will have to create time averaged MD structures myself because the average structures (pdb files) which I had from the Plin folder are not the one which we are really interested.
Will keep you posted once I am done with this. A revise draft will all revision requested will follow subsequently.
Vijayan_task_list1_RC_comments_3-19_2016_1.docx
RC (3/19): Please see attached. The revisions requested should be the immediate focus and continue for the whole week if needed.
Some essential points require clarification. Please provide brief replies regarding questions on Mon where possible prior to addressing the requests in more detail.
Vijayan_task_list1_RC_comments_3-19.docx
If time permits in between or thereafter, you can list the simulation parameters from the identified input files under each simulation used in Figs/Tables in the paper.
RSK: 3/18/2016
RSK: Document attached below is a clean version ( excluding the comments/communications which we exchanged during the course of revision ) of tasklist_1 with the final version of the fig/fig legends/ tables.
Task_list1_final_cleaned.docx
RSK:3/17/2016
RSK: I have uploaded the revised version of Tasklist_1.
I have ensured that all the comments which had suggested on the wiki page are also copied in to the document.
The questions and comments which you had for me are highlighted in RED, my responses to them are highlighted in GREEN and any remark which I had are highlighted in Cyan.
The document also contains the old and the final revision of the figures.
In case if you feel that you want to have a look at the final revised figures/legends/plots, in the interest of time, then let me know. I can send you a cleaned up document which only contains the final version of the figs and plots and fig legends.
Task_list1_final_revised.docx
XG(3.16.2016)
Mutation Table_02.04.2015.docx
Sequence alignment_02.09.2015.pdf
RSK: I have revised the figure appropriately. All other revision which you had suggested will be carried out tomorrow (work on progress).
RC: Further revisions will be required thereafter.
Please make sure all my earlier comments have been incorporated in the document (the comments themselves).
It may take a day for me to review your changes. Please take your time to make the revisions properly. After you have completed the next set of revisions,
propose your next steps that you will work on while waiting for feedback.
RSK: Okay, will ensure that all your comments are addressed.
The new figure is inserted at page 10 of the revised document . let me know if its okay or needs another round of revision. I have used the right structures.
Another version without the substrate will be added tomorrow.
Task_list_R3_1Revisions.docx
3/14/2016
RSK: Will revise it accordingly as per your latest comments and upload a revised version shortly.
3/14/2016
Revisions to Task_list 1
RSK: I have completed the revisions that you had asked for.
The figs have been recreated, and labelled. Captions have been added and also have been revised.
RC: I'll have to take a closer look at the MD average fig - it was not immediately clear to me. Are you still working on it?
Regarding Ping's comment on C vs B pocket / Phe conformation, yes he made a comment about some difference in Phe conformation
between INT and INT:NAM complexes. It wasn't clear from his comment how significant this conformational difference was.
Please provide a general status update as well (remaining tasks).
RC:
- I don't think all my earlier comments regarding the tasks are incorporated in the doc (e.g., I don't see all the comments
regarding the MD average fig).
- I think you have access to all the files on the wiki, but I have posted in dropbox a ppt created by Ping on loop conformations,
given their relevance to some of these figures.
RSK: I do have access to Plin's Dropbox folder. Will check it.
RC: I meant that I put the required ppt in the For Vijayan dropbox.
- Regarding the B/C pocket issue for Phe: I noted that the different figs provide varying perspectives on the active site.
It may be easier to observe the difference in Phe conformation between the INT complex (shown in green in
Figure XXXXX: Superposition of Sirt3 native intermediate) and in INT:NAM complex if the figures used the same perspective.
RSK: Will ensure that figures have the same perspective in the revised version.
- Regarding the sequence alignments and annotations of catalytically important residues in other figures: please get the
table from Guan on the roles of these residues and include some version of this at the end of the document so it is self-contained
and so the captions can later be revised to include some mention of the roles of these residues if desired.
This should also be available on the wiki.
RSK: Okay, will revise it.
Regarding caption for sequence alignment, I didn't see a draft of the condensed/revised version of the original caption that Ping
apparently borrowed from the sequence alignment program.
- I am not sure that we are on the same page regarding the MD average figure. See the proposed caption,
which mentions that the two loop conformations after MD are to be compared.
I only see a single purple loop in the figure provided. Is that the INT or ternary conformation of the loop?
Perhaps this is related to the potential issue you had referred to in a previous posting.
This figure needs more work.
The latter is probably the most important pending issue.
RSK: Will get it done by today.
In this figure, I'm not sure that using different colors for the loops adds to clarity. Perhaps a different rendering or other approach
could be considered.
RSK: Okay, will remove the colors for the loop
What structures did you use for the latest version of this figure (R2 revision)? The intention was to use the MD averages from 4FVT simulations with and without loop replacement from
4BVG. Is that what you used or did you use a simulation based on 4BVG protein structure for the intermediate?
RSK:
In the revised figure I used an MD average structure (last 10ps) of 4FVT_isoNAM.pdb ( 4FVT-IsoNAM- with loop modelled from 4BVG) superimposed onto native 4FVT structure
I think that I will have to revise this figure using the 4FVT having NAM instead of iso NAM ( ie, 4FVT-NAM- with loop modeled from 4BVG), because I see that is what you require. I think that now we are on the same page on the how the figure should be created. If not let please me know.
In the figure based on pdb structures, you showed the substrate. In this case would it compromise clarity?
RSK: You mean the Acy-Lys. I will create another one without out the substrate
RC: By "In this case" I meant the MD average figure. It is lacking the substrates.
RSK: Okay got it. Will create two version w/wo substrate. Will eventually use the one with better clarity.
Regarding B/C pocket, it does appear from the latest version of the figure that Phe is closer to the C pocket for the INT loop conformation compared to ternary,
but not in the C pocket as in the case of 4BVG (INT complex). In these simulations NAM is in the C pocket.
The above details of the structure preparation (loop substitution, presence of NAM) need to be clear in caption (see original version of caption).
RSK: Okay, got it.
If you have any questions regarding the details of this proposed figure please let me know.
RSK: I think i got your point. I will get you a new figure that has 4FVT-NAM-loop region modlled for 4BVG superimposed onto a MD avergae structure of native 4FVT. Also, I will ensure that the loops are not differently colored form the rest.
Task_list_R1_Revisions.docx
Progress status of my Task_list 1
Task_list1_Status.docx
RC: Thanks. Please add my most recent comments, including indication of which (if any) figures had draft captions
included in the task list, and the comments regarding the MD average figure.
Regarding task 2, we may have the structures match those that will be included in the MD average Fig. We could start with that, following
which I will consider further and advise.
For task 2, you mentioned we can easily edit displays for alternate versions.
Zooming in closer to the loop may be desirable in one version.
Are we still doing a version with side chains displayed?
Please see feedback in the following revision:
Task_list_R2.docx
Task_list_R1.docx